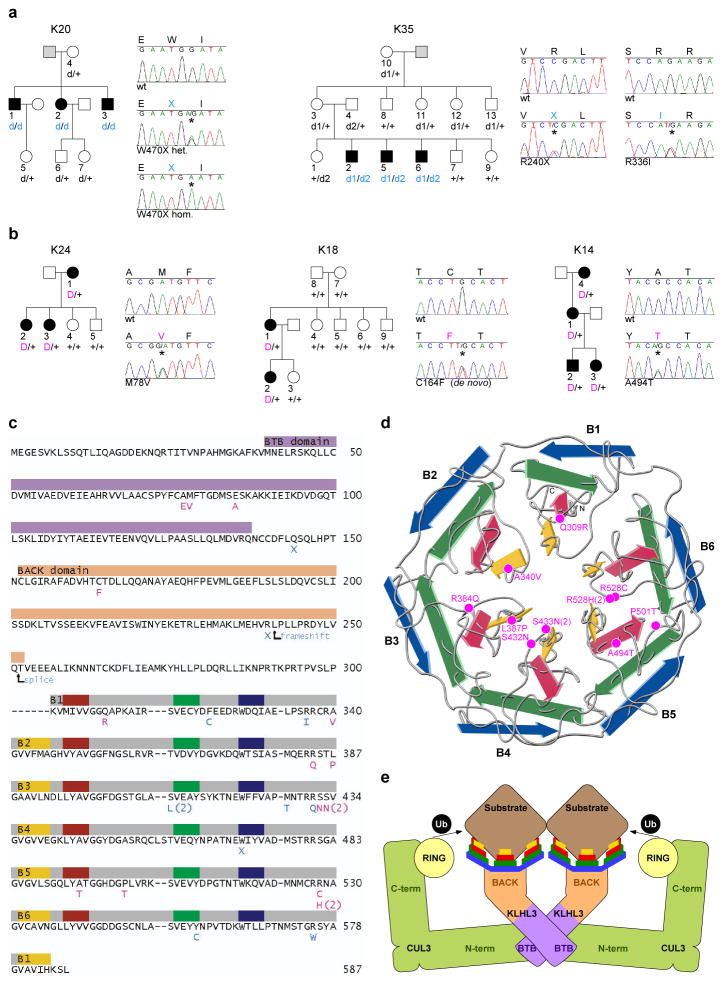
Figure 1. Recessive and dominant KLHL3 mutations in PHAII kindreds.
a–b, Representative kindreds demonstrating recessive (a) and dominant (b) KLHL3 mutations (all 24 kindreds are shown in Supplementary Figs. 3–4). Affected, unaffected, and phenotype-undetermined subjects are denoted by black, white, and gray symbols, respectively. KLHL3 alleles are denoted by ‘+’ (wild-type), ‘d’ (recessive mutation), and ‘D’ (dominant mutation). Sequence traces show wild-type (wt) and mutant (*) alleles and encoded amino acids. c, KLHL3 protein sequence. Colored bars indicate BTB domain (lavender), BACK domain (peach), and Kelch propeller blades (B1-B6, gray) with β-strands ‘a’–‘d’ in yellow, red, green, and blue respectively. Recessive (aqua) and dominant (pink) mutations are shown; recurrences indicated by numbers. d, Kelch propeller schematic, from KLHL2 crystal structure27. β-strands colored as in c; dominant mutations indicated. e, CRL schematic, comprising a BTB-Kelch protein (KLHL3), CUL3, and a ubiquitin transfer-mediating RING protein, with substrate bound via the Kelch propeller. Complex shown as a dimer7.