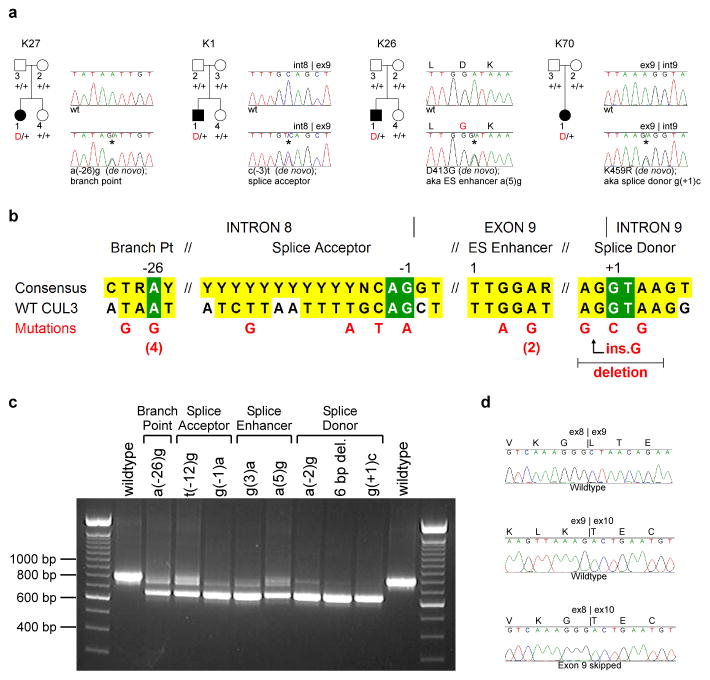
Figure 2. Dominant CUL3 mutations in PHAII kindreds cause skipping of exon 9.
a, Representative kindreds demonstrating CUL3 mutations, depicted as in Fig. 1 (all 17 kindreds are shown in Supplementary Fig. 6). b, CUL3 mutation locations. Consensus splicing sequences18,28 and corresponding wildtype CUL3 sequences within intron 8, exon 9, and intron 9 are shown; invariant bases (green) and consensus homology (yellow) are indicated. Positions numbered relative to splice sites and first base of the exonic splice (ES) enhancer. Mutations shown in red; recurrences indicated by numbers. c, RT-PCR of spliced RNA. Wild-type CUL3 constructs produce a single product including exons 8, 9, and 10 (844 bp); all nine mutants tested produce a predominant product that skips exon 9 (673 bp). d, Representative RT-PCR sequences. Wild-type construct produces cDNA with properly spliced junctions between exons 8–9 (top) and 9–10 (middle), while mutant construct [splice donor g(+1)c] produces cDNA joining exon 8 to exon 10 (bottom).