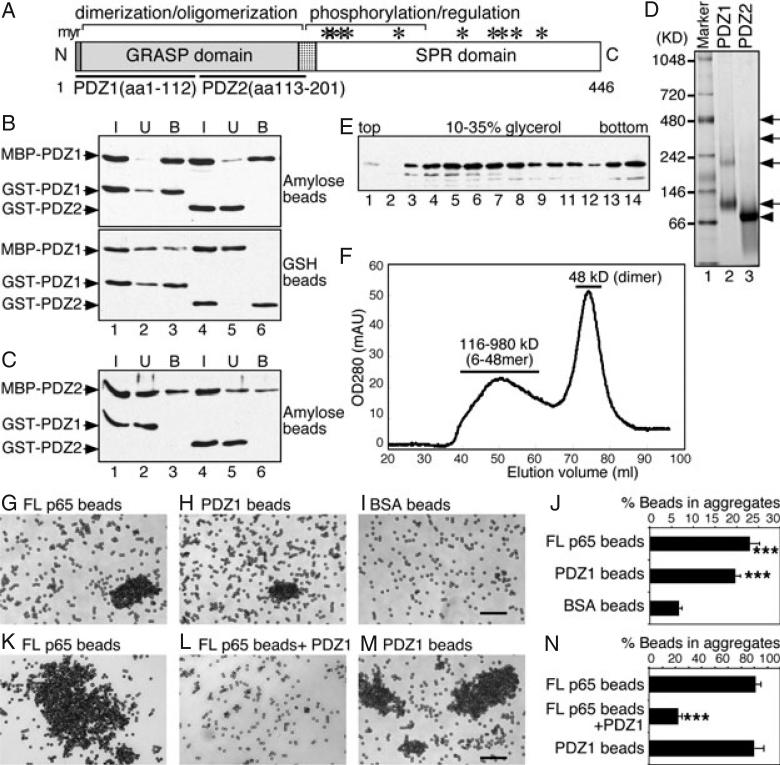
Figure 1. The N-terminal half of the GRASP domain (PDZ1) is sufficient to form dimers and oligomers.
A) Domain structure of GRASP65. Indicated are the N-terminal GRASP domain with myristoylated N-terminal glycine (myr) and the C-terminal serine/proline-rich (SPR) domain with phosphorylation sites (*). The GRASP domain contains two PDZ subdomains, PDZ1 (aa 1–112) and PDZ2 (aa 113–201). B) PDZ1 is sufficient to form oligomers as shown by a copurification assay. Separately purified recombinant MBP-tagged PDZ1 was mixed and incubated with GST-tagged PDZ1 or PDZ2. The protein complexes were isolated using either amylose or glutathione (GSH) beads, as indicated. Equal proportions of the input (I), unbound (U) or bound (B) fractions were analyzed by immunoblotting for GRASP65. Note that PDZ1 copurified with itself (lane 3), but not with PDZ2 (lane 6). C) PDZ2 does not form oligomers. As in (B) but using MBP-PDZ2 as the bait. Note that PDZ2 did not copurify with PDZ1 (lane 3) or itself (lane 6). D) Analysis of oligomerization of PDZ1 and PDZ2 by non-denaturing gels. GST-tagged PDZ1 and PDZ2 were incubated at 37°C and analyzed by non-denaturing electrophoresis and stained with Coomassie Blue. Molecular weight standards are indicated on the left. Note that PDZ1 formed dimers and oligomers (arrows), while PDZ2 formed dimers (arrowhead) only. E) Analysis of PDZ1 oligomerization by velocity gradient. Purified recombinant MBP-tagged PDZ1 was sedimented in 10–35% glycerol gradients. Shown is the western blot for GRASP65 in each fraction. PDZ1 is found in both the upper (4–7; dimers) and deep fractions (13–14; higher-order structures). F) Gel filtration analysis of PDZ1. Purified His-tagged PDZ1 was analyzed by gel filtration using a Sephacryl S-300 column. The protein was separated into two peaks: a broad peak of 116–980 kDa (6-48mer) and a sharp peak of 48 kDa (dimer). Sizes were calculated according to the calibration with a gel filtration molecular weight standard (Sigma). G–I) Bead aggregation assay showing that PDZ1 is sufficient to form oligomers. Beads coated with His-tagged full-length (FL) GRASP65 (G), PDZ1 (H) or BSA (I) were incubated with BSA at 37°C for 60 min. After incubation, the beads were placed on glass slides and random fields were photographed. A representative image of each condition is shown. Bar, 50 μm. Note that FL and PDZ1-coated beads formed aggregates. J) Quantitation of the percentage of beads in aggregates in (G–I) from three independent experiments. ***p < 0.001, compared with BSA-coated beads. K–M) Bead aggregation assay showing that soluble PDZ1 inhibits oligomerization of FL GRASP65. Beads coated with His-tagged FL GRASP65 (K and L) or PDZ1 (M) were incubated with interphase cytosol at 37°C for 60 min. In (L), purified recombinant PDZ1 was added into the reaction. After incubation, the beads were placed on glass slides and random fields were photographed. A representative image of each condition is shown. Bar, 50 μm. Note that PDZ1 can inhibit the aggregation of GRASP65-coated beads. N) Quantitation of the percentage of beads in aggregates in (K–M). Results in (J) and (N) expressed as the mean ± SEM from three sets of independent experiments. ***p < 0.001, compared with FL GRASP65-coated beads in the absence of PDZ1 added to the reaction.