Abstract
This paper describes the AEO, an ontology of anatomical entities that expands the common anatomy reference ontology (CARO) and whose major novel feature is a type hierarchy of ~160 anatomical terms. The breadth of the AEO is wider than CARO as it includes both developmental and gender-specific classes, while the granularity of the AEO terms is at a level adequate to classify simple-tissues (~70 classes) characterized by their containing a predominantly single cell-type. For convenience and to facilitate interoperability, the AEO contains an abbreviated version of the ontology of cell-types (~100 classes) that is linked to these simple-tissue types. The AEO was initially based on an analysis of a broad range of animal anatomy ontologies and then upgraded as it was used to classify the ~2500 concepts in a new version of the ontology of human developmental anatomy (www.obofoundry.org/), a process that led to significant improvements in its structure and content, albeit with a possible focus on mammalian embryos. The AEO is intended to provide the formal classification expected in contemporary ontologies as well as capturing knowledge about anatomical structures not currently included in anatomical ontologies. The AEO may thus be useful in increasing the amount of tissue and cell-type knowledge in other anatomy ontologies, facilitating annotation of tissues that share common features, and enabling interoperability across anatomy ontologies. The AEO can be downloaded from http://www.obofoundry.org/.
Keywords: anatomical hierarchy, cell-type assignations, ontology, tissue classification
Introduction
Formal anatomical ontologies are now an important component of the informatics infrastructure of model organism and other databases (Bard, 2008; for a review of anatomy ontologies, see the papers in Burger et al., 2008; for examples, see1) and are also a key part of the informatics tools intended to explore biomedical databases. These ontologies primarily use part_of as their main structural relationship (e.g., every heart is part_of a cardiovascular system) because the smaller anatomical entities (usually referred to as tissues) are naturally seen as the constituent parts of larger ones, albeit that one tissue may be a part of more than one anatomical system (e.g., the femur is part_of the lower limb and the skeletal system). In addition, this relation is particularly important within database schemas for querying such tissue-associated knowledge as gene-expression data (e.g., the totality of the genes expressed in the heart at some developmental stage is the sum of the genes expressed in its parts).
In addition to part_of relationships, anatomical ontologies also need a classification or type hierarchy in which every term is related by an is_a or type relationship to a higher class term (e.g., the femur is_a bone, the deltoid is_a muscle). This relationship is required for three reasons: first, to ground the ontology within a standard formal structure (ontologies are based on classes within superclasses); second, many ontology visualization tools require this relationship; and third, this classification assigns to a term anatomical knowledge that would otherwise be missing.
An informal way of handling this issue is to indicate tissue type within an anatomy ontology through the use of high-level terms (e.g., leg skeleton, limb muscle system, cranial ganglia) but, while this is sometimes adequate for navigation around the ontology, it cannot be viewed as satisfactory or rigorous because it is based on a part_of rather than an is_a or type relationship. A better approach has been to use the common anatomy reference ontology (CARO) to classify anatomical structures (Haendel et al., 2008). This very high-level ontology of anatomical types is intended to provide a coarse framework of low-granularity for referencing the tissues of adult organism on the basis of anatomical level. Its 80 or so terms cover all anatomical classes from a hermaphroditic organism to an epithelium’s basal lamina; only about 16 of them, however, can be used to classify tissues and cell-types (e.g., organ system, compound organ, multi-tissue structure). The only histological classification in the CARO covers the different types of epithelia; no other tissues (e.g., neuronal, muscular, and mesenchymal) merit a mention.
While the CARO provides a high-level class for a structure of any scale and so can be used to satisfy the requirement that every class have a superclass, its very low-granularity means that it can only annotate the thousands of tissue types that are known with very limited knowledge about anatomical structure. The restrictions of the CARO have been informally discussed within the field for some time and additions are beginning to be made. Thus the curator of the Drosophila anatomy ontology needed to add a few new type terms (e.g., row) for classifying adult fly tissues. More recently, the vertebrate musculoskeletal anatomy ontology (namespace: VAO) has been produced (see text footnote 1), also using the CARO for its high-level terms, and this ontology meets the need for a new and much richer set of classes for this subset of anatomy. A more serious omission in CARO is that, because it was designed for adult anatomies, it lacks terms for developing tissues, a major focus of many anatomical ontologies. These and other class terms have been included in Uberon (Washington et al., 2009), an integrated cross-species ontology with high-level CARO terms and classified by structure, function, and developmental lineage, but not in any detail by tissue type. It is thus clear that an ontology for anatomy tissues that is both richer and finer-grained than CARO is required if one wishes to include structural knowledge about tissues in anatomy ontologies.
This paper describes the ontology of anatomical entities (AEO), an expansion of the CARO. The AEO is intended to capture and classify knowledge about anatomical structures not currently included in anatomical ontologies and includes ~100 new classes structured using the is_a relationship. The AEO terms were selected partly through analysis of histology and anatomy books, partly through logical analysis, partly for their use in classifying the new ontology of human developmental anatomy (~2500 terms) and partly through examination of a range of animal ontologies (whose ids are included where appropriate). The granularity of the AEO terms is at a level adequate for tissues of a predominantly single cell-type and, and these are given through has_part relationships to an abbreviated version of the ontology of cell-types (~90 classes) included in the AEO. The AEO may be useful in increasing the amount of tissue and cell-type knowledge in other anatomy ontologies, facilitating annotation of tissues that share common features, and enabling interoperability across anatomy ontologies.
Materials and Methods
The AEO uses the CARO as its basis for high-level classes. Terms for the histological information used to link cell-types to tissues came from standard textbooks (e.g., Ross et al., 1995; Standring, 2008; human anatomy is, for obvious reasons, analyzed in far greater depth than that of other organisms). Additional terms came from an analysis of other adult and anatomical ontologies from the biomedical ontologies site, particularly the VAO and, in these cases, the original ids are stored as dbxrefs. All ontologies mentioned in the paper are available from the OBO foundry (see text footnote 1). In this context, it might have been appropriate to incorporate within the AEO the terms and the structure of the VAO. The major skeletal terms from the VAO have been included (with definitions and ids), but the structure of the VAO was not used, mainly because it is much larger, more complex, and more fine-grained than is appropriate for the AEO and partly because some of the finer details of classification is at odds with expectation.
The process of constructing the AEO is described below. In brief, a first draft was made on the basis of inspection of a wide range of anatomical ontologies combined with general reading. This was used to classify the ontology of human developmental anatomy which has ~2500 concepts. This process exposed weaknesses and omissions that were successively corrected.
Because the granularity of the AEO is designed to include anatomical entities of a single cell-type (simple-tissue or its synonym portion of tissue), it seemed sensible to include these cell-types within the ontology. While this could have been done using dbxrefs to the cell-type ontology, it seemed more appropriate to include the cell-type terms within the ontology so that a partonomy relationship could be assigned. A subset of the cell-type ontology was therefore included within the AEO and its terms linked to appropriate simple-tissue via the has_part relationship which carries the meaning that tissue A includes within it at least some of cell-type B.
The AEO terms not originally present in the CARO carry AEO ids whose numbers do not overlap with CARO ids (see Discussion) and is authored in the obo format2 using the OBO-Edit3 (Day-Richter et al., 2007.) and CoBrA4 (Aitken et al., 2005) browsers (the former for complex ontologies, the latter for simple ones). Terms also carry appropriate dbxrefs from the Drosophila, VAO, zebrafish, Uberon, and human developmental anatomy ontologies. Obo-Edit includes the ability to make disjoint_from links that facilitate inconsistency checking (Rector, 2003) and such links have been made for male and female anatomical structures, and for material and immaterial anatomical structures.
The obo ontology is available from the OBO foundry (see text footnote 1). For Protégé users, the OWL version is generated automatically by the OBO Foundry pipeline, and is available from the same URL.
Results
Design features
The key aim of the AEO was to provide at least one unambiguous type term for every tissue in the anatomical ontology for an animal, whether adult, or developing. This turned out to be a more complicated process than originally expected, and what is described below is the final result of a series of iterations as drafts of the AEO were used for annotation (see below). The initial stage in making the AEO involved making a series of choices.
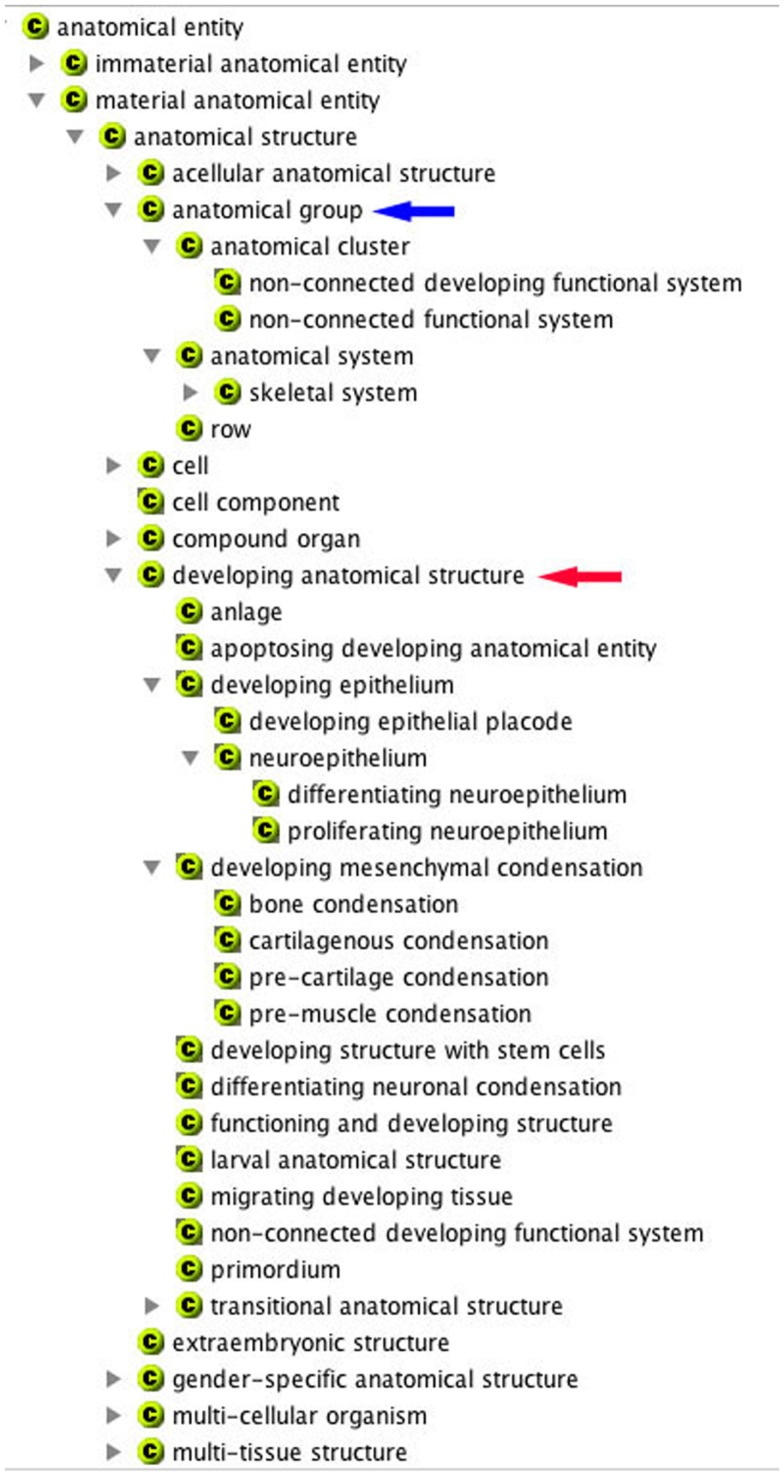
The first decision resulted from considering whether further high-level terms were needed in the CARO, and two omissions were noted: the exclusion of gender-specific and embryonic anatomical entities. The former was straightforward to add, but the latter, important for anatomy ontologies that cover developing organisms, was more difficult. The problem in choosing subterms here lies in the fact that all tissues in an embryo are developing tissues (even if they are fully functional and just growing, e.g., the late metanephros) and there is little point in annotating every term in an ontology with is_a developing tissue. As a result, a minimalist view was taken here and the terms in the developing tissue branch of the ontology were limited to those that were likely to be populated, were not present in an adult organism and had a useful developmental implication (Figures 1 and 2). Excluded from the list are any terms that imply lineage (such as may be found in Uberon); this is mainly because there are few if any tight lineage restrictions on tissue morphology. The list is of developmental classes is thus short and may need to be extended.
Figure 1.
The AEO shown in the COBrA browser. Here, the hierarchies for immaterial anatomical entities (blue arrow) and gender-specific anatomical entities (red arrow) are expanded.
Figure 2.
The AEO shown in the in the COBrA browser. Here, the hierarchies for anatomical group (blue arrow) and developing anatomical structure (red arrow) are expanded.
The second decision focused on the depth of the ontology, and here a CARO definition proved key: the CARO defines a portion of tissue as “anatomical structure that consists of similar cells and intercellular matrix, aggregated according to genetically determined spatial relationships.” This definition fits comfortably with an anatomist’s view of the simplest tissue by implying that it has a defined boundary and has cells predominantly of a single class (although this definition does raise the occasional problem – see below). One advantage of going down to this simple level of structure was that it enabled each leaf term to be annotated with its cell-types (as detailed in the cell-type ontology).
The third decision in making the AEO lay in choosing the breadth of the hierarchy. The coverage should be good enough to be useful without being overwhelmingly detailed, and anatomists have produced very detailed catalogs of tissue classes: Gray’s anatomy (Standring, 2008), for example, lists >8 types of joint, most of which are rare. In making the AEO, all the major animal ontologies (i.e., plant and fungal ontologies are excluded) available at the OBO library (see text footnote 1) were examined, and terms were chosen on the basis that they were likely to be useful (i.e., populated) and clear in meaning to anatomists. Thus, only the two most common classes of joint (synovial and fibrous joints) are included in the AEO as specific subclasses of joint; the former is_a multi-tissue structure and the latter is_a a simple-tissue, while cartilage is not subdivided. Also excluded are accessory bones (a subclass of sesamoid bones), bursas (a subclass of epithelial sac), and venules and arterioles (because they are all both unnamed and dispersed). Because skeletal terms are so common and useful to anatomists and to evolutionary biologists, it seemed sensible to group them all as parts under a new term skeletal system, a subclass of anatomical system.
There is a further small point: the terms of the AEO are intended to be clear in meaning to any biologist: as the ontology is intended for experimentalists who wish to annotate terms and access data, it is therefore important that the terms be those in common use. In this context, no anatomist has an intuitive sense of what the CARO term portion of tissue means, so the AEO uses that tem as a synonym for its replacement simple-tissue (similarly, the term portion of organism substance has been made a synonym of the more intuitively obvious term non-tissue substance).
Making the ontology
The major structural additions to the CARO that seemed necessary beyond adding developmental and gender-specific tissues were the expansion of some top-level class terms such as immaterial anatomical entity, anatomical groups, and organism subdivision, and here it seemed sensible to include the obvious major categories (head, body, etc., see below). Similarly, the class multi-tissue structure was felt to be too broad and in need of subterms, perhaps the most important of which is tissues with stem cells.
The class of immaterial anatomical entities (i.e., terms that refer to features rather than tissues) is treated lightly in the CARO: its few terms merely specify dimension (anatomical line, point, surface, and space). This terseness does less than justice to the richness of surfaces and volumes in organisms so the AEO includes several more terms (Figure 1) that can be used to group immaterial entities with common topological features (e.g., open and enclosed cavities, Figure 1). One interesting question here concerned how to class surface pits and grooves (e.g., the otic pit): should they be viewed as anatomical spaces (3D) or as surface features? Perhaps the most logical way to handle this would be to view the cells bounding the feature as a simple-tissue and the enclosed space (with a virtual enclosing surface) as an immaterial anatomic space. This would mean distinguishing between, say, the otic pit space and the otic pit epithelium, but standard anatomical usage implies that the otic pit is actually a surface feature within the surface epithelium. After some thought, the latter option was chosen with the user having the further option of annotating the term with a tissue type, so allowing both the cell-type and the geometric feature to be captured. Should a user specifically wish to refer to the space within the pit, the volume can be classified as a lumen of an epithelial sac. There is, it should be said, some vagueness in saying that an entity can be both a material and an immaterial entity; the values in doing this are terseness and the ability to captures some sense of tissue geometry, the price is the risk, albeit small, of ambiguity.
The other key task was the choice of simple-tissue leaf terms and this was mainly done on the basis of analyzing anatomy ontologies and histology texts. The net result was a major expansion in the CARO class simple-tissue (portion of tissue) which now has eight subclasses rather than one, with these subclasses opening up to two further levels which cover a further 60 or so classes (Figure 3). One anomalous term that has been included under neuronal tissue is nerve fiber tract: even though such tracts are composed of axons rather than of complete cells and so are not a tissue in the normal meaning of the word, this term was included because nerve fiber tracts are both named and important. As neither the CARO nor the cell-type ontology has a natural class that includes anatomical entities composed of cell parts, the GO definition for neuron projection bundle (and GO id dbxref) has been used here (and the synonym included). In a sense, all neuronal tissues are anomalous because the cell bodies and axons are not found within the same structure and it would have seemed odd to have included nerve fiber tract under any heading other than simple-tissue.
Figure 3.
The simple-tissue hierarchy of the AEO shown in the COBrA browser. All top and secondary levels terms are shown together with some tertiary level ones.
As a result of this, a draft extension to the CARO was constructed with ~70 new terms.
Improving draft versions of the AEO
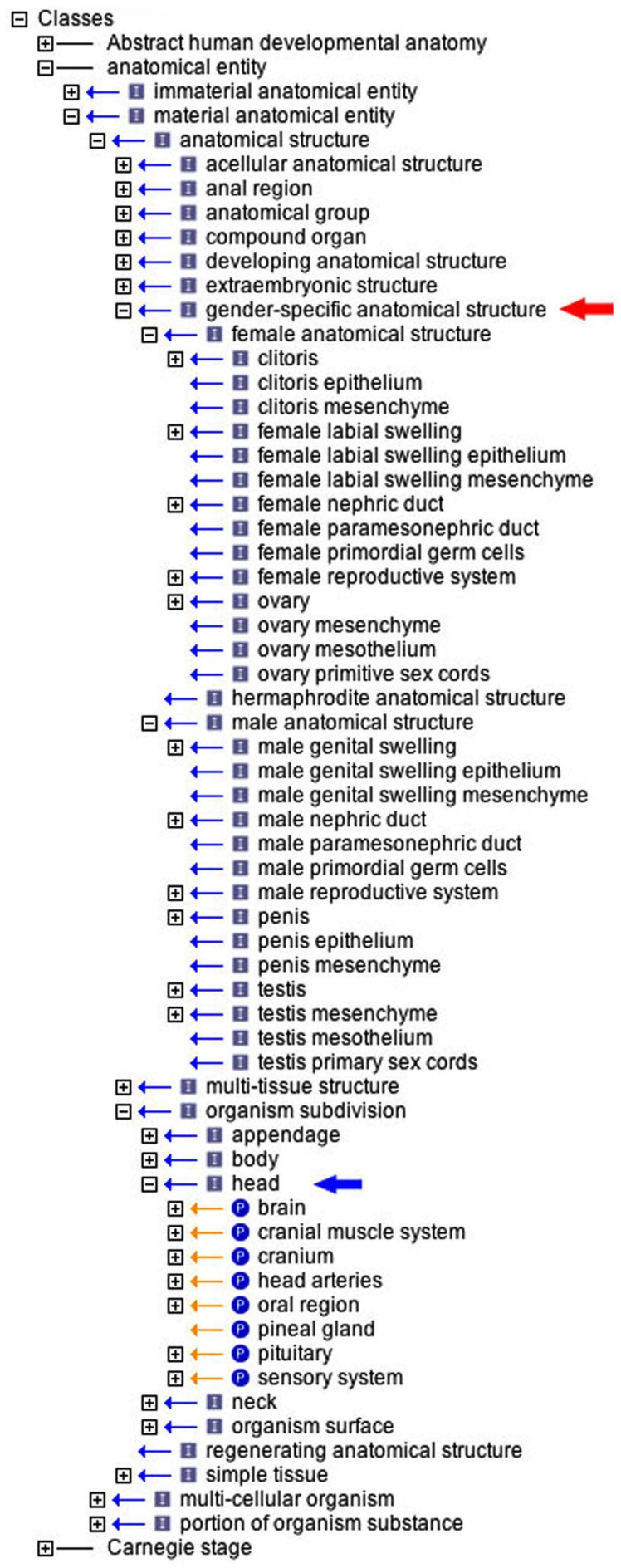
The AEO is intended to provide an is_a link for any anatomical concept. As the initial draft was based on inspection of a range of anatomical ontologies for animals, it met this criterion for most animal tissues. A harsher and finer granularity test was its ability to provide type terms for all the concepts in a detailed anatomical ontology. For this, drafts of the AEO were used to provide an obvious type term for the ~2500 tissues in the new and integrated ontology of human developmental anatomy (namespace: EHDAA2; current draft available from http://www.obofoundry.org/) which is currently being constructed by the author from one made a decade ago (Hunter et al., 2003) that included a separate ontology for each Carnegie stage (1–20). The process of annotating a very wide range of anatomical classes from major organ systems down to simple-tissues in EHDAA2 identified inadequacies in draft AEO ontologies and required many changes to both the terms and the structure of the AEO. The introduction of developing anatomical structure and gender-specific embryological structure has already been mentioned (Figures 1 and 2). Another example was the amplification of organism subdivision. This last category proved useful, for example, in grouping the many and disparate entities within the head using part_of relationships (Figure 5). As things currently stand, there is at least one easily assignable class term for all anatomical terms so far examined, be it a leaf node (e.g., metanephric mesenchyme is_a developing mesenchymal condensation) or a higher level concept (somite group is_a row).
Figure 5.
The cell-type hierarchy of the AEO shown in the COBrA browser. This subset of the full cell-type ontology includes about 90 classes in all but only the epithelial cell class is expanded (red arrow). The editor uses the has_part relationship to show those anatomical entities (simple-tissues) which include the various epithelial cell-types.
During this exercise, another ~30 terms were added. In the current version, 13 of the new terms are classes of immaterial anatomical entity, ~20 are developing anatomical entities, ~40 are new types of simple-tissue, 15 are multi-tissue structures, seven are anatomical groups, and a few others are distributed under various headings. Further terms can easily be added if users feel that they would be needed. The current version of the AEO thus includes ~160 anatomical classes and ~100 cell-types.
Adding cell-type relationships
Once the AEO was in place, it seemed sensible to supplement the anatomical type information by annotating the leaf tissues of simple-tissue and the appropriate subclasses of multi-tissue structure with their cell-types and these are formally detailed in the cell-type ontology. This ontology is unnecessarily rich for the fairly simple annotation exercise here and a sub-ontology of the ~100 cell-type classes that were needed (~15% of the original ontology) was made on the basis of standard histology textbooks and incorporated within the AEO (Figure 4). There is no ideal relationship to convey the sense that a particular cell-type is a major but not exclusive constituent in a particular class of tissue (there may be several cell-types in such relatively simple-tissues as ganglia, epithelia, and mesenchymal domains, Table 1). The relationship chosen for the link was has_part and this carries the meaning that tissue A has some part made of cell-type B, as can be seen by inspection of the Obo file (Table 1). Unfortunately, browsers require that the relationships be read upward and so the link is under the cell-type rather than under the tissue (Figure 5).
Figure 4.
The use of the AEO in classifying the ontology of human developmental anatomy (EHDAA2). The ontology is opened in the OBO-Edit browser to demonstrate (i) those tissues classed (is_a relationship) within the gender-specific anatomical structure hierarchy (red arrow), and (ii) the head category of organism subdivision with its constituent organ groups (part_of relationship).
Table 1.
The AEO obo file entry for autonomic ganglion.
| (Term) |
|---|
| ID: AEO:0001001 |
| Name: autonomic ganglion |
| Namespace: anatomical_entity_ontology |
| Def: “a ganglion that is part of the autonomic nervous system.” (JB:AEO) |
| Is_a: AEO:0000135! ganglion |
| Relationship: has_part CL:0000107! autonomic neuron |
| Relationship: has_part CL:0000243! glial cell (sensu vertebrata) |
| Relationship: has_part CL:0000526! afferent neuron |
| Relationship: has_part CL:0000527! efferent neuron |
| Relationship: has_part CL:0002573! Schwann cell |
Making the cell-type to simple-tissue relationships was usually straightforward, in the sense that one cell-type could usually be seen as the predominant type for a tissue, but this was not always so, particularly for the tissues of the nervous system such as ganglions where neurons are always accompanied by support cells.
Discussion
There were two key reasons for producing the AEO: first, to provide a formal type definition for the ontology of human developmental anatomy so that it would meet modern ontology standards and, second, to enable this and perhaps other anatomy ontologies, which are mainly built from part_of relationships, to increase the amount of anatomical knowledge that they contain. This new ontology had, of course, to be based on the CARO scaffold, as its use is now standard for anatomy ontologies.
In practice, the only problem in using the CARO as a scaffold turned out to derive from the definition of the term simple-tissue (or portion of tissue) whose definition was “anatomical structure, that consists of similar cells and intercellular matrix, aggregated according to genetically determined spatial relationships.” This definition assumes that a single structure (the anatomical term) is composed of essentially similar cells, and, while this is usually so, there are some important exceptions, particularly in the nervous system where ganglia and brain nuclei where neurons, the key functional cell-type in the nervous system: neurons in ganglia and brain nuclei are always accompanied by support cells (glia, astrocytes, etc.). Another type of structure that could, on the basis of its boundaries, be viewed as simple is the membrane bone which includes osteoblasts, osteoclasts, and osteocytes. Placing such structures within the AEO could not be done in any natural way, and the solution adopted was based on what seemed to be the most appropriate location for a user. Membrane bone was made a subclass of bone, a multi-tissue structure, while ganglion and neuronal nucleus were made subclasses of neuronal structure, a simple-tissue. While these choices are not logically consistent, it is to be hoped that they will not lead to any downstream problems.
Attaching the cell-type terms to the new classes was done relatively late in the production of the AEO as it became clear that it would be quite simple and straightforward to add them from a subset of the cell-type ontology. Although there is always the concern that a user might suppose that the relationships are complete, they are not! It should be emphasized that only key cell-types have been included. Thus, for example, most tissues in an organism have associated blood vessels, nerve endings, and phagocytic cells and these have not been included. The relationship used here is has_part (every endochondral bone has_part one or more osteocytes) and this allows the cell-types to be associated with the tissues in the obo file. Browsers views this relationship in an inverted way and show the tissues associated with a cell-type (Figure 5).
A key part of making the ontology was using its classes to annotate the anatomical terms in the ontology of human developmental anatomy (EHDAA2) being revised from that of Hunter et al. (2003). This annotation exercise frequently demanded that new AEO terms be added and occasionally that their location be changed. As a result of annotating the EHDAA2 ontology and glancing through other vertebrate ontologies the terms seem adequate for typing vertebrate tissues. The same amount of attention has not, it should be said, been given to invertebrate ontologies and those working in this area may well find that, if they choose to use the AEO, they will need additions or changes (see below).
The only reason for producing a new ontology is that it should be useful and I hope that the integration of AEO within anatomy ontologies other than that for human developmental anatomy may prove helpful in two contexts at least. First, it would help curators who wish to annotate and users who wish to search on the basis of anatomical structure (e.g., all ducted glands). Second, it would be of value to anyone who wishes to know something about the histology of a tissue and the sort of cells that it contains. In this context, the simple-tissue hierarchy may be particularly useful to both groups. In addition, the AEO can rightly be seen as no more than an expansion in breadth and granularity of the CARO and it is a fair question as to whether the AEO should absorbed within the CARO with AEO ids becoming CARO ids. This is really a question that the curators of the CARO and of anatomy ontologies other than that for human developmental anatomy will need to answer; if they do decide to do this, the transfer will be easy: as there is no current overlap in id number between the CARO and AEO namespaces, it will merely require a global change in the AEO OBO file of <AEO:> to <CARO:> (provided, of course, that no new terms are added to CARO in the meantime).
The ontology is named the Anatomical Entity Ontology and this might seem a little ambitious, given that its focus is primarily on vertebrate and secondarily on invertebrate anatomy, with little attention so far being paid to plants and fungi anatomy. In practice, there are terms within the AEO that can be used to type such tissues (e.g., nectar and sap are non-tissue substance, cambium, and root meristem are developing tissues with stem cells, a dictyostelium slug is_a migrating tissue and hyphae is_a epithelial plexus). That said, the AEO does not yet contain specific plant and fungal terms and it is intended that future drafts will include appropriate type term for classifying organisms from these phyla. It is also planned that they will include semantic features automating classification (Rector, 2003; Meehan et al., 2011).
Although drafts of the AEO have been discussed with others (see Acknowledgments), the ontology will inevitably have errors and omissions. Suggestions, comments and criticisms should be sent to j.bard@ed.ac.uk. This and further versions of the ontology will be posted at and downloadable from the OBO foundry http://www.obofoundry.org/. A summary of the ontology can be found at http://www.obofoundry.org/wiki/index.php/AEO:Main_Page and this wiki will be used to post details of future changes.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Richard Baldock, Albert Burger, Duncan Davidson, Melissa Haendel, Gillian Morriss-Kay, and David-Osumi-Sutherland for discussion. I am grateful to Chris Mungall for always being available for email conversations, for commenting on the manuscript, regularizing the relationships, and for maintaining the obo site. I also thank the reviewers for their helpful criticisms.
Footnotes
References
- Aitken S., Korf R., Webber B., Bard J. (2005). COBrA: a bio-ontology editor. Bioinformatics 21, 825–826 10.1093/bioinformatics/bti097 [DOI] [PubMed] [Google Scholar]
- Bard J. B. L. (2008). “Anatomical ontologies for model animals,” in Anatomy Ontologies for Bioinformatics, eds Burger A., Davidson D., Baldock R. (Springer: ), 3–25 [Google Scholar]
- Burger A., Davidson D., Baldock R. (eds). (2008). Anatomy Ontologies for Bioinformatics. New York: Springer [Google Scholar]
- Day-Richter J., Harris M. A., Haendel M., The Gene Ontology OBO-Edit Working Group. Lewis S. (2007). Obo-Edit – an ontology editor for biologists. Bioinformatics 23, 2198–2200 10.1093/bioinformatics/btm112 [DOI] [PubMed] [Google Scholar]
- Haendel M. A., Neuhaus F., Osumi-Sutherland D., Mabee P., Mejino J. L. V., Mungall C., Smith B. (2008). “CARO, the common anatomy reference ontology,” in Anatomy Ontologies for Bioinformatics, eds Burger A., Davidson D., Baldock R. (New York: Springer; ), 327–349 [Google Scholar]
- Hunter A., Kaufman M. H., McKay A., Baldock R., Simmen M. W., Bard J. B. L. (2003). An ontology of human developmental anatomy. J. Anat. 203, 347–355 10.1046/j.1469-7580.2003.00224.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan T. F., Masci A. M., Abdulla A., Cowell L. G., Blake J. A., Mungall C. J., Diehl A. D. (2011). Logical development of the cell ontology. BMC Bioinformatics 12, 6. 10.1186/1471-2105-12-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A. (2003). Modularisation of domain ontologies implemented in description logics and related formalisms including OWL. Proc. KCAP ACM 2003, 121–129 [Google Scholar]
- Ross M. H., Romrell L. J., Kaye G. I. (1995). Histology: a Text and Atlas. Baltimore: Williams & Wilkins [Google Scholar]
- Standring S. (ed.). (2008). Gray’s Anatomy, 40th Edn London: Churchill Livingstone [Google Scholar]
- Washington N. L., Haendel M. A., Mungall C. J., Ashburner M., Westerfield M., Lewis S. E. (2009). Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 7, e1000247. 10.1371/journal.pbio.1000247 [DOI] [PMC free article] [PubMed] [Google Scholar]