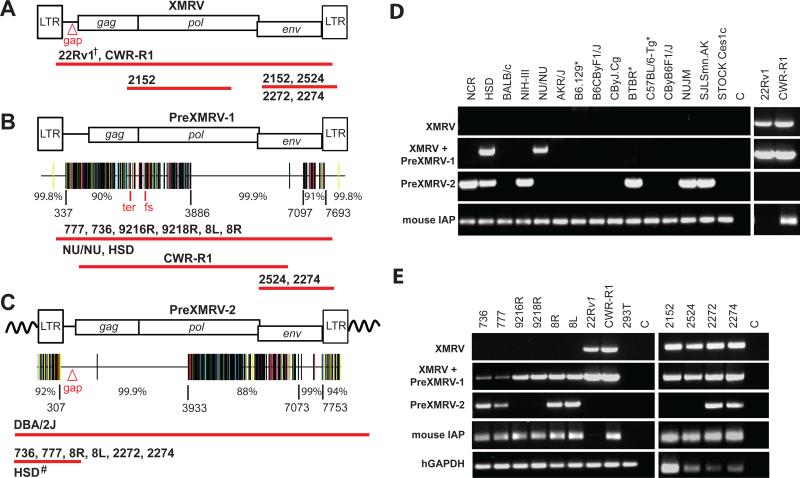
Fig. 2.
PCR and sequencing analysis of XMRV and XMRV-related sequences from xenografts, cell lines, and nude mouse strains. Using specific primer sets (Fig. S2), cloned PCR products from the xenografts, 22Rv1, CWR-R1, or mouse strains were sequenced. Approximate length and location of sequences determined from samples that were positive for XMRV (A), PreXMRV-1 (B) and PreXMRV-2 (C) are shown as red bars beneath each provirus. Details of primers and numbers of cloned products sequenced are shown in Figs. S2 and S3. Hypermut plots (see Fig. S3 for details), which indicate nucleotide mismatches relative to XMRV as color-coded vertical lines, are shown for PreXMRV-1 (B) and PreXMRV-2 (C), together with the percent identity to consensus XMRV for different regions of each provirus (nucleotide numbers refer to the 22Rv1 XMRV sequence [FN692043]). PreXMRV-1 has a 16-nt deletion (Δ16) in gag and a frameshift (fs) in pol making it replication defective while PreXMRV-2 gag, pol, and env reading frames are open. Mouse strains (D) and xenograft and PC cell lines (E) were analyzed by PCR for the presence of XMRV, PreXMRV-1 and PreXMRV-2. Mouse IAP and human GAPDH serve as positive controls for the presence of mouse and human DNA, respectively. For both (D) and (E) the primer set used to detect PreXMRV-1 can also detect XMRV. For ease of comparison, the 22Rv1 and CWR-R1 gel lanes from (E), which were run in parallel, are duplicated in (D). DNAs in (D) and (E) were all amplified with the same PCR primer master mix. †We previously determined the full-length sequence of XMRV from 22Rv1 cells (8). Δgap refers to the 24-bp deletion in the gag leader characteristic of XMRV. All mouse strains shown in (D) are nudes except for those indicated with *.