ETOC: Two aspects of cohesin's diverse functions are its regulation by acetylation and its contributions to cell viability. When Smc3p acetylation is constitutive or absent, little if any cohesion is established. Cells remain viable because Smc3p acetylation and Wpl1p regulate chromosome condensation, and a cohesin-independent segregation exists.
Abstract
Cohesin generates cohesion between sister chromatids, which enables chromosomes to form bipolar attachments to the mitotic spindle and segregate. Cohesin also functions in chromosome condensation, transcriptional regulation, and DNA damage repair. Here we analyze the role of acetylation in modulating cohesin functions and how it affects budding yeast viability. Previous studies show that cohesion establishment requires Eco1p-mediated acetylation of the cohesin subunit Smc3p at residue K113. Smc3p acetylation was proposed to promote establishment by merely relieving Wpl1p inhibition because deletion of WPL1 bypasses the lethality of an ECO1 deletion (eco1Δ wpl1Δ). We find that little, if any, cohesion is established in eco1Δ wpl1Δ cells, indicating that Eco1p performs a function beyond antagonizing Wpl1p. Cohesion also fails to be established when SMC3 acetyl-mimics (K113Q or K112R,K113Q) are the sole functional SMC3s in cells. These results suggest that Smc3p acetylation levels affect establishment. It is remarkable that, despite their severe cohesion defect, eco1Δ wpl1Δ and smc3-K112R,K113Q strains are viable because a cohesin-independent mechanism enables bipolar attachment and segregation. This alternative mechanism is insufficient for smc3-K113Q strain viability. Smc3-K113Q is defective for condensation, whereas eco1Δ wpl1Δ and smc3-K112R,K113Q strains are competent for condensation. We suggest that Smc3p acetylation and Wpl1p antagonistically regulate cohesin's essential role in condensation.
INTRODUCTION
An evolutionarily conserved “cohesin” complex physically tethers sister chromatids (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998; Sumara et al., 2000; Tomonaga et al., 2000). In budding yeast, the cohesin subunits are called SMC3, SMC1, MCD1/SCC1, and SCC3/IRR1 (Guacci et al., 1997; Michaelis et al., 1997). Sister chromatid cohesion is established during S phase and maintained until anaphase, when sister chromatids segregate. Inactivation of cohesin induces precocious dissociation of sister chromatids and cell inviability (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998; Tomonaga et al., 2000; Vass et al., 2003). These results led to the conclusion that sister chromatid cohesion is essential for the bipolar attachment of sister kinetochores, proper chromosome segregation, and, consequently, cell viability in all eukaryotes.
In addition to its role in cohesion, cohesin also plays roles in mediating proper mitotic and meiotic chromosome condensation in many eukaryotes (Guacci et al., 1997; Revenkova et al., 2004; Ding et al., 2006; Jin et al., 2009), in transcription regulation (Donze et al., 1999; Dorsett, 2007; Schaaf et al., 2009), and in repair of double-stranded DNA breaks (Strom et al., 2007; Unal et al., 2007). These additional cohesin functions raise two questions. How are these different functions of cohesin regulated, and do any of these other functions contribute to cell viability? The cohesin regulator Eco1p (also called Ctf7p) provides a common link since it functions in budding yeast chromosome condensation, as well as cohesion (Skibbens et al., 1999)
Cohesin has been most extensively studied in its role in mediating cohesion. Its binding to chromosomes is spatially and temporally regulated. Cohesin is found at discrete arm loci called cohesin-associated regions and at pericentric regions (Onn et al., 2008). In budding yeast, cohesin binds chromosomes during late G1 phase in a form that cannot establish cohesion, and then during S phase, Eco1p converts cohesin into a “cohesive” form capable of generating cohesion (Skibbens et al., 1999; Toth et al., 1999). Insights into the biochemical basis for this conversion came from the identification of Eco1p as an acetyltransferase whose substrates include several cohesin subunits (Ivanov et al., 2002; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Onn et al., 2009). The key Eco1p target is Smc3p, which is acetylated at two evolutionarily conserved lysine residues, which are K112 and K113 in budding yeast (Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). K113 is the critical residue, as replacement with arginine, a similar but nonacetylatable residue (K113R), blocks cohesion generation and cell viability, whereas the K112R mutation has no effect (Unal et al., 2008; Zhang et al., 2008). Moreover, the cohesion defect of an eco1 temperature-sensitive (ts) mutant is significantly reduced by addition of an allele of SMC3 with an acetyl-mimic glutamine at K113 (K113Q), whereas the smc3-K112Q allele has no effect (Unal et al., 2008). However, the K113Q allele has little to no ability to restore viability to the eco1 mutant (Unal et al., 2008). This raised the possibility that the acetyl-mimic has some limitation or that Eco1p is required for an essential cohesin function distinct from cohesion.
Recent work provided genetic clues into the function of Eco1p-dependent acetylation. Whereas ECO1 is an essential gene in budding and fission yeasts, certain mutations in the cohesin regulators WPL1 (also called RAD61) or PDS5 enabled strains deleted for ECO1 (eco1Δ) to be viable (Tanaka et al., 2000; Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009; Feytout et al., 2011). In mammalian cells, Wapl1p (the vertebrate Wpl1p orthologue) appears to antagonize cohesin, as its depletion stabilizes cohesion, whereas its overexpression induces cohesion loss (Gandhi et al., 2006; Kueng et al., 2006). Together these observations led to the model in which Wpl1p inhibits cohesin to block establishment and Smc3p-K113 acetylation antagonizes Wpl1p during S phase, thereby eliminating the inhibition and allowing cohesion establishment. However, budding yeast eco1Δ wpl1Δ double mutants have a severe chromosomal arm cohesion defect when assayed in M phase–arrested cells (Rowland et al., 2009; Sutani et al., 2009). Owing to the dogma that cohesin's role in cohesion is the essential function, the eco1Δ wpl1Δ cell viability was assumed to reflect proper establishment of cohesion but subsequent failure to maintain it through M phase (Rowland et al., 2009; Sutani et al., 2009). It is important to note that this assumption was never tested. Moreover, cohesion at centromeric proximal loci was never tested, making it possible that centromeric cohesion was normal in eco1Δ wpl1Δ cells and thereby promoted segregation. Cohesion at both CEN-proximal and distal loci must be more rigorously characterized to elucidate how Wpl1p modulates cohesion establishment in budding yeast.
In summary, previous studies of Eco1p regulation of cohesin yielded two surprising observations that uncouple levels of cohesion and cell viability. First, Smc3p acetyl-mimics integrated into an eco1 ts mutant cannot support viability despite their ability to significantly restore arm cohesion. Second, eco1Δ wpl1Δ cells are viable despite their severe arm cohesion defect. These apparently paradoxical results prompted us to reexamine cohesion, chromosome structure, and viability in Smc3p acetyl-mimic–bearing cells and in eco1Δ wpl1Δ cells. Our results provide important and surprising insights into the roles of Eco1p, Smc3p acetylation, and Wpl1p in cohesin regulation both for cohesion and its noncohesion functions.
RESULTS
The smc3-RQ allele, but not the K113Q allele, supports cell viability
We previously showed that an smc3 acetyl-mimic K113Q allele integrated into an eco1-ts mutant significantly suppressed the eco1-ts cohesion defect but had little or no ability to suppress its inviability (Unal et al., 2008). Because the eco1-ts allele is severely compromised for its acetyltransferase activity, we suggested that one possible reason for the partial cohesion restoration by the acetyl mimic and its failure to promote viability was that Eco1p acetylation of other targets might be required (Onn et al., 2009; Unal et al., 2008). In addition, the eco1-ts mutant contained an endogenous WT SMC3 allele, which could potentially compete with the acetyl-mimic and limit its effects. We eliminated these issues by integrating acetyl-mimic alleles into an smc3-42 ts mutant. First, this strain bears an endogenous wild-type ECO1 gene. Second, at nonpermissive temperature, the mutant smc3-42p cannot assemble cohesin or bind chromosomes, but the smc3 acetyl-mimics assemble cohesin (Toth et al., 1999; Unal et al., 2008; Sutani et al., 2009). Therefore, at nonpermissive temperature, we assay Smc3p acetyl-mimics when they comprise the sole functional cohesin in the presence of Eco1p activity.
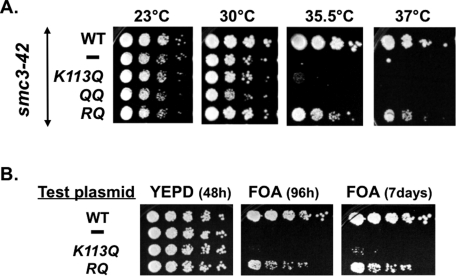
We first tested the ability of smc3 acetyl-mimic alleles to suppress the smc3-42 mutant temperature sensitivity. Cells grown at permissive temperature were dilution plated onto yeast extract/peptone/dextrose (YPD) media and then incubated at different temperatures to assess viability (see Materials and Methods). As expected, the parent smc3-42 ts mutant was unable to grow at either 35.5 or 37°C, whereas the integrated wild-type (WT) SMC3 enabled growth at all temperatures (Figure 1A). Neither the smc3-K113Q nor the K112Q,K113Q (QQ) allele enabled growth at 35.5 or 37°C. In contrast, an smc3-K112R,K113Q (RQ) allele–bearing strain grew relatively well at both 35.5 and 37°C.
FIGURE 1:
The smc3-RQ allele promotes viability but the K113Q or QQ alleles do not. (A) Effect of smc3 acetyl-mimics on smc3-42 temperature sensitivity. Haploid VG3358 3B (smc3-42) alone (–) or with a second SMC3 allele, (WT) VG3377-1A, (K113Q) VG3423-1A, (QQ) VG3378-2A, or (RQ) VG3424-2A, grown at 23°C, plated at 10-fold serial dilutions on YPD, and incubated 3 d at 23, 30, 35.5, or 37°C. (B) Ability of smc3 acetyl-mimic alleles to support viability as the sole SMC3. Haploid VG3464-16C with pEU42 (SMC3 CEN URA3) and test plasmid bearing either no insert (–), SMC3 (WT), smc3K113Q (K113Q), or smc3K112R,K113Q (RQ) grown at 30°C, plated in fivefold serial dilutions on FOA or YPD, and incubated at 30°C.
To confirm the difference between the K113Q and RQ alleles with regard to supporting viability, we assessed their function in the absence of any other SMC3 allele, using the shuffle strategy. We began with a strain deleted for the chromosomal copy of SMC3 but kept alive by the presence of plasmid pEU42 (SMC3 URA3 CEN). Cells were transformed with a second “test” plasmid (CEN LEU2) bearing wild-type SMC3, the K113Q allele, the RQ allele, or no insert. Strains were tested for the ability to survive with only the test plasmid by growing cells on media containing 5-fluoroorotic acid (5-FOA), which selectively kills URA3+ cells, thereby selecting for cells that have lost pEU42 (see Materials and Methods). As expected, cells bearing the wild-type SMC3 test plasmid grew well on FOA, whereas cells bearing the empty vector failed to grow (Figure 1B). The K113Q test plasmid also failed to enable growth on FOA. In contrast, the RQ test plasmid allowed growth on FOA, albeit with slower growth rates and increased inviability than wild-type SMC3 (Figure 1B). These results confirm that the smc3-RQ allele can support viability, whereas the K113Q allele cannot. Because the K112 residue can be acetylated in the K113Q allele but not the RQ allele, our results suggest that hyperacetylation in the K112, K113 region is detrimental to cohesin function.
Mutants defective in cohesin subunits and cohesin regulators can be sensitive to benomyl, a microtubule inhibitor, and to camptothecin, a topoisomerase I inhibitor that induces DNA damage (Aguilar et al., 2005; Heidinger-Pauli et al., 2010a). Indeed, strains bearing the RQ allele in an smc3-42 background or as the sole SMC3 allele were sensitive to these drugs, as well as being cold sensitive (Supplemental Figure S1, A–C). Therefore, even though the RQ allele enables viability, it is compromised for some aspect(s) of cohesin function.
The smc3-RQ allele promotes viability despite being severely defective in cohesion establishment
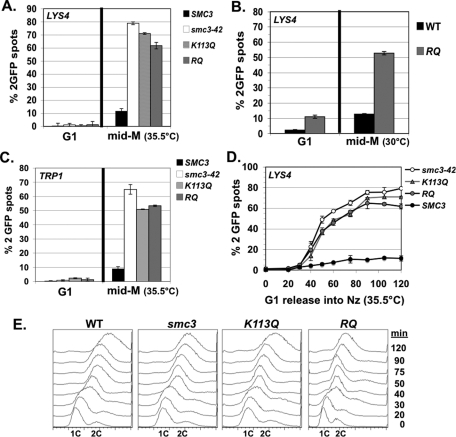
We next investigated the mechanism by which the smc3-RQ allele supports viability but the K113Q allele does not. Given the importance of sister chromatid cohesion, we assayed cohesion in a parent smc3-42 strain alone or bearing an additional integrated SMC3 allele—the wild type, the K113Q, or the RQ allele. Mid-log-phase cells were arrested in G1 phase (using α factor) at permissive temperature (23°C) for smc3-42 and then synchronously released at the nonpermissive temperature (35.5°C) in the presence of nocodazole to rearrest in mid M phase. G1 and mid M phase–arrested cells were scored for cohesion at LYS4 (located 470 kb from CEN4 on the chromosome IV right arm) using a LacO/LacI system and for DNA content using flow cytometry (see Materials and Methods).
As expected, nearly 80% of the parent smc3-42 haploid cells had two green fluorescent protein (GFP) spots, indicating a severe cohesion defect, whereas adding WT SMC3 generated normal cohesion, so that few cells had two GFP spots (Figure 2A). For both the K113Q- and RQ-bearing strains, >60% of cells had two GFP spots, indicating that neither allele efficiently restored cohesion. G1-phase cells in all strains had a single GFP spot, demonstrating that mid M-phase cells with two GFP spots were not due to preexisting aneuploidy. We next assayed cohesion in strains in which either WT SMC3 or the smc3-RQ alleles provide the sole SMC3 source. Cells were synchronously released from G1 phase (using α factor) and then rearrested in mid M phase using nocodazole (see Materials and Methods). Cohesion was assayed at the arm (LYS4) locus (Figure 2B). As expected, few mid-M-phase WT cells had two GFP spots. In contrast, the smc3-RQ allele cells had two GFP spots in >50% of mid M phase–arrested cells, indicating defective arm cohesion and confirming results for the RQ allele in the smc3-42 strain.
FIGURE 2:
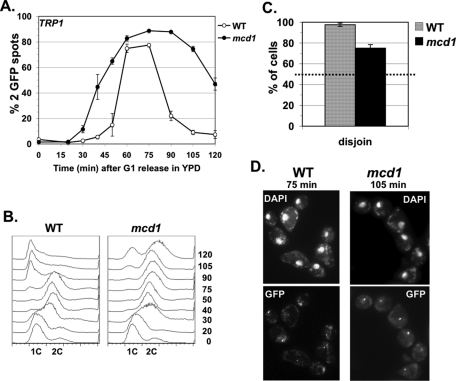
smc3 acetyl-mimics are defective in cohesion at CEN-proximal and distal loci. Cells released from G1 and then arrested in mid M phase. The percentage of cells with two GFP signals is plotted. (A) Cohesion loss at CEN-distal locus (LYS4) in smc3-42 background at mid M phase (35.5°C). Haploid VG3358-3B (smc3-42) alone or with a second SMC3 allele, (WT) VG3377-1A, (K113Q) VG3423-1A, or (RQ) VG3424-2A. (B) Cohesion loss at CEN-distal locus (LYS4) in shuffle strains at mid M phase (30°C). Strains have SMC3 (WT) VG3471-WT or (RQ) VG3471-WT as sole SMC3. (C) Cohesion loss at a CEN-proximal locus (TRP1) in smc3-42 background at mid M phase (35.5°C). Haploid VG3357-3A (smc3-42) alone or with a second SMC3 allele, (WT) VG3447-1B, (K113Q) VG3449-3B, or (RQ) VG3450-4B. (D) Time course to monitor cohesion loss at a CEN-distal locus (LYS4). Strains from A scored at 10- to 15-min intervals after G1 release into mid-M-phase arrest (35.5°C). (E) DNA content. Data were derived from two independent experiments, and error bars are SD. For cohesion assays, 100–400 cells were scored for each data point in each experiment.
A simple explanation for viability of smc3-RQ bearing cells is that cohesion forms at CEN-proximal regions to enable bipolar spindle attachments. To assess this possibility, we examined cohesion at a centromere-proximal locus (TRP1) located 10 kb from CEN4 (see Materials and Methods). Haploid smc3-42 ts cells alone or also bearing a WT or smc3 acetyl-mimic were synchronously arrested in mid M phase at 35°C, using nocodazole as described. Similar to the arm locus, mid-M-phase cells from the smc3-RQ- or K113Q-allele bearing strains were defective for cohesion, whereas cohesion was restored by wild-type SMC3 (Figure 2C). Thus the smc3-RQ and K113Q alleles are defective in their ability to generate cohesion at both CEN-proximal and distal loci.
Mutants in the cohesin-associated Pds5p establish cohesion at nearly wild-type levels but fail to maintain it during mitosis (Tanaka et al., 2001; Stead et al., 2003). Therefore we performed a time course to assess whether the cohesion loss observed in mid M phase–arrested smc3-RQ or K113Q cells was due to a defect in cohesion establishment or maintenance. Cells were synchronously released from G1 arrest and then rearrested in mid M phase at 35.5°C, using nocodazole as described, except that cell aliquots were taken at 10- to 15-min intervals following release from G1 phase. As expected for the WT SMC3 allele, few cells had two GFP spots at any time point (Figure 2D). The smc3-42 mutant alone or bearing either the smc3-RQ or K113Q allele had two GFP spots arising near the time of replication, indicating a defect in cohesion establishment (Figure 2D). Together these data show that both the K113Q and RQ alleles have the same severe defect in the establishment of cohesion.
The eco1Δ wpl1Δ double mutant is viable but has a severe defect in cohesion establishment
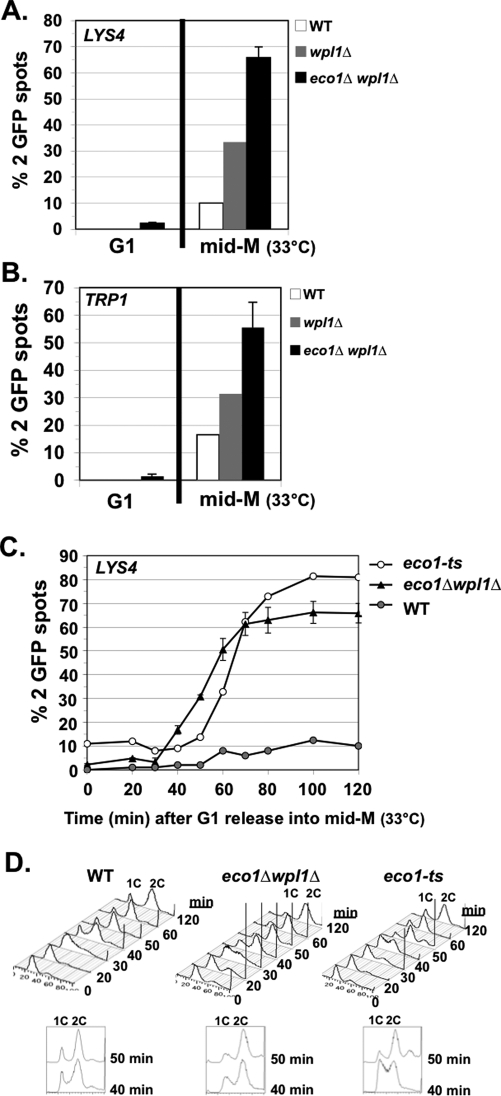
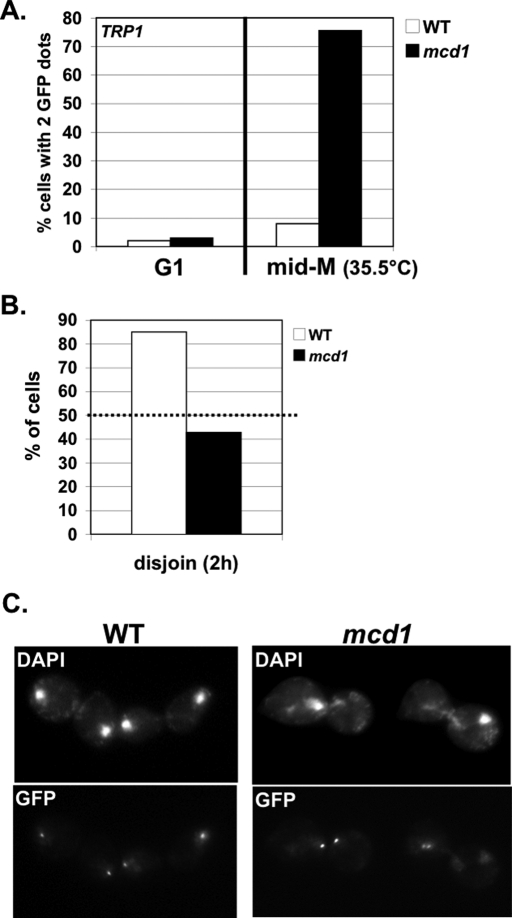
Recent studies suggested that Smc3p acetylation merely serves to remove Wpl1p-mediated inhibition to cohesion establishment, because budding yeast cells deleted for both ECO1 and WPL1 (eco1Δ wpl1Δ) are viable (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009). Our data showing that the smc3-RQ or K113Q acetyl-mimics fail to establish cohesion is inconsistent with this simple model since the mimics should antagonize Wpl1p. However, one cannot exclude the possibility that the acetyl-mimics are not functionally equivalent to Smc3p acetylation. Therefore we assayed cohesion at the LYS4 arm locus in eco1Δ wpl1Δ double mutants synchronously arrested in mid M phase, using nocodazole as described. For controls, we assayed wild-type cells and a wpl1Δ single mutant. Wild-type cells have cohesion, so very few cells had two GFP signals (Figure 3A). Most eco1Δ wpl1Δ cells had two GFP spots, indicating a major cohesion defect, whereas the wpl1Δ single mutant had a moderate defect (Figure 3A). Such a major defect in arm cohesion was previously seen in eco1Δ wpl1Δ cells arrested in mid M phase (Rowland et al., 2009; Sutani et al., 2009). We also assayed cohesion at the centromere-proximal locus (TRP1) in cells synchronously arrested in mid M phase. Most eco1Δ wpl1Δ cells had two GFP spots, indicating a major cohesion defect (Figure 3B). Thus eco1Δ wpl1Δ cells are defective for cohesion at both CEN-proximal and distal loci.
FIGURE 3:
eco1Δ wpl1Δ cells are defective in cohesion at CEN-proximal and distal loci. The percentage of cells with two GFP signals is plotted. Cells released from G1 phase were arrested in mid M phase at 33°C. (A) Cohesion loss at a CEN-distal locus (LYS4) in mid M phase. Haploids VG3349-1B (WT), VG3360-3D (wpl1Δ), VG3503-1B (eco1Δ wpl1Δ), and VG3503-4A (eco1Δ wpl1Δ). (B) Cohesion loss at CEN-proximal locus (TRP1) in mid M phase. Haploids VG3460-2A (WT), VG3513-1B (wpl1Δ), VG3502-2A (eco1Δ wpl1Δ), and VG3503-4C (eco1Δ wpl1Δ). (C) Kinetics of cohesion loss at a CEN-distal locus (LYS4). Haploids VG3349-1B (WT), VG3506-5D (eco1-ts), VG3503-1B (eco1Δwpl1Δ), and VG3503-4A (eco1Δ wpl1Δ) assayed for cohesion loss after release from G1 phase. (D) FACS. Data were derived from two independent experiments, and error bars are SD. For cohesion assays,100–400 cells were scored for each data point in each experiment.
We next performed a time course to determine when cohesion was lost at the LYS4 arm locus in eco1Δ wpl1Δ cells. WT and eco1-ts (eco1-203; originally called ctf7-203) cells were used as controls. Cells were synchronously released from G1 phase into media containing nocodazole at 33°C, the nonpermissive temperature for the eco1-ts allele. Aliquots were analyzed for cohesion and DNA content at 10- to 15-min intervals. Very few wild-type cells had two GFP spots, whereas 70% of cells in both the eco1Δ wpl1Δ and eco1-ts mutants had two GFP spots. More important, cohesion loss appeared concomitant with DNA replication in both eco1Δ wpl1Δ and eco1-ts mutants (Figure 3, C and D). Of interest, there was ∼10-min delay in DNA replication in eco1-ts cells compared with eco1Δ wpl1Δ cells (compare 40-min time points), which corresponded to ∼10-min delay in the appearance of two GFP spots in the eco1-ts cells (Figure 3, C and D). Small perturbations in replication have been noted when cohesin or cohesin regulators are perturbed, but the mechanism responsible for this phenomenon is unclear (Skibbens, 2011). Finally, we assayed eco1Δ wpl1Δ cells for sensitivity to benomyl and camptothecin. The eco1Δ wpl1Δ cells are highly sensitive to both drugs and exhibit increased inviability compared with WT or wpl1Δ cells (Supplemental Figure S2). This dual sensitivity and lower viability is similar to that of the smc3-RQ cells and is consistent with a loss of cohesion (Supplemental Figure S1, A and B). Thus eco1Δ wpl1Δ cells fail to establish cohesion, and this, together with data from cells bearing the smc3-RQ allele, supports our conclusion that budding yeast cells can survive with extremely poor cohesion establishment.
Sister chromatids segregate despite severe defects in cohesion establishment
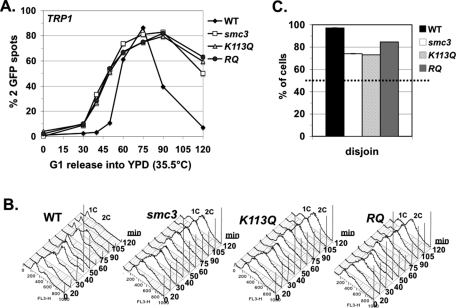
Our results raise two important questions. First, if cohesion is essential for bipolar attachments and sister chromatid segregation, how can the smc3-RQ and the eco1Δ wpl1Δ strains be viable with such poor cohesion establishment? Second, given that they exhibit the same large cohesion defect, why are smc3-RQ allele–bearing and eco1Δ wpl1Δ mutant strains viable, whereas the K113Q allele–bearing strain is inviable? Because sister chromatids must segregate to opposite poles to enable viability, we examined whether the smc3-RQ allele can somehow promote segregation in smc3-42 cells, whereas the K113Q allele cannot. Strains were synchronously released from G1 phase at 35.5°C into YPD media but, unlike before, without adding nocodazole. This regimen allowed spindle formation, chromosome segregation, and cell division. We readded α factor as soon as buds formed in most cells to allow completion of only one cell cycle and subsequent rearrest in G1 phase. Cells were marked at the CEN4-proximal (TRP1) locus. We scored the number of GFP spots per cell and their position within cells to assay cohesion and segregation, respectively (see Materials and Methods).
We first analyzed when sister chromatid cohesion was lost. Wild-type cells had only one GFP signal from G1 phase though completion of S phase, indicative of normal cohesion (Figure 4, A and B). WT cells entered anaphase by 60 min, and so sister cohesion is dissolved and most cells have two GFP signals. This percentage decreased from 90 min onward as chromosome segregation is completed and cells exit mitosis and rearrest in G1 phase (Figure 4, A and B). In contrast, for the smc3-42 mutant, alone or with either the K113Q or the RQ allele, cells with two GFP signals began accumulating from the time of replication (Figure 4, A and B). Therefore establishment is defective at a CEN-proximal locus, as well as at the arm locus. Cells accumulated as large-budded cells with 2C DNA content (Figure 4, A and B), indicative of delay in mitosis, which is expected for cells defective in cohesion (Guacci et al., 1997).
FIGURE 4:
Sister chromatids segregate in smc3-42 mutants with or without acetyl mimics. Strains from Figure 2C smc3-42 mutant (smc3) or with a second SMC3 allele, WT, K113Q, or RQ, released from G1 at 35.5°C to allow one cell cycle before rearrest in G1. Chromosome IV monitored at a CEN4-proximal locus (TRP1). (A, B) Time course assessing cohesion loss. (A) Percentage of cells with two GFP signals. (B) DNA content. (C). Chromosome IV sister chromatid disjunction scored in large-budded cells. Dotted line marks the 50% disjunction expected for random segregation. For cohesion and nondisjunction assays, 100–400 cells were scored for each data point in each experiment.
We next analyzed sister chromatid disjunction in anaphase/telophase cells. Chromosomes were scored as having disjoined when one GFP spot (sister chromatid) segregated into each cell body of a large-budded cell. Sister chromatids were scored as exhibiting nondisjunction when two GFP spots cosegregated into one cell body, whereas the other had no GFP spot (see Materials and Methods). Because cohesion enables bipolar attachments, cells with defective cohesion should be unable to achieve bipolar attachments, and random (50% disjunction) segregation of sister chromatids is expected. For WT cells, anaphase/telophase cells were most abundant at 75 min after release, and so disjunction was scored at that time. As expected, disjunction was observed in virtually all large-budded cells (Figure 4C). The two cell bodies contained equal-sized 4′,6-diamidino-2-phenylindole (DAPI) masses, consistent with disjunction of all sister chromatids (data not shown). The RQ, K113Q, and parent smc3-42 mutants were scored at 120 min after release since most remained in anaphase/telophase cells, and only some cells completed division (Figure 4C). It is surprising that, despite the failure to establish cohesion, disjunction was observed in 75% or more of smc3-42 mutants cells alone or containing the K113Q or the smc3-RQ allele (Figure 4C). Moreover, the two cell bodies often contained two equal-sized DAPI masses, consistent with disjunction of most sister chromatids. We also scored the smaller number of G1 phase cells that exited mitosis (see Materials and Methods). The results were similar to that seen with large-budded cells—sister chromatids disjoined in most cells (Supplemental Figure S3A). Thus sister chromatids disjoin surprisingly well even when cohesion establishment is dramatically impaired.
The nonrandom segregation of sister chromatids observed with smc3-42 mutants suggests budding yeast does not require cohesin-mediated cohesion to promote segregation. To test this conclusion further, we performed the same disjunction assay using a mutant in the MCD1 cohesin subunit (mcd1-1), previously shown to be defective for establishment (Guacci et al., 1997; Noble et al., 2006). As before, WT cells had one GFP signal from G1 phase until anaphase onset, when cells with two GFP spots accumulated, and then decreased as cells exit mitosis and rearrest in G1 phase (Figure 5, A and B). WT anaphase/telophase cells show disjunction in almost all cells by 75 min, with equal-sized and completely separated DNA masses present in each bud (Figure 5, C and D). The mcd1-1 mutant cells, like the smc3-42 mutant cells, had two GFP signals during and soon after replication due to a failure to establish cohesion, and then accumulated as large-budded cells with two GFP signals (Figure 5, A and B). By 105 min, disjunction was observed in >75% of anaphase/telophase mcd1-1 cells (Figure 5C). Moreover, two equal-sized separated DNA masses were seen, consistent with disjunction of most sister chromatids (Figure 5D). Similar high levels of disjunction were observed in unbudded (G1 phase) mcd1-1 cells that had completed one cell cycle (Supplemental Figure S3B). We noted that segregation was delayed in mcd1-1 cells, as shown by comparison of the 75-min time points in mcd1-1 and wild-type cells, but ultimately sister chromatids disjoined (Supplemental Figure S3C and Figure 5D). Therefore, in mcd1-1 and smc3-42 mutant cells, most sisters precociously dissociate but subsequently properly disjoin, revealing the existence of a cohesin-independent segregation pathway. This cohesin-independent pathway helps to explain how the smc3-RQ strain and likely the eco1Δ wapl1Δ strain are viable despite their dramatic defect in cohesion establishment.
FIGURE 5:
Sister chromatids segregate in mcd1-1 mutant cells released from G1 phase at nonpermissive temperature. Haploid WT (VG3460-2A) and mcd1-1 mutant (VG3456-2C) cells released from G1 arrest at 35.5°C and allowed to complete one cell cycle. Chromosome IV was monitored at a CEN4-proximal locus (TRP1). (A, B) Time course to assess cohesion loss. (A) Percentage of cells with two GFP signals. (B) DNA content. (C). Chromosome IV sister chromatid disjunction scored in large-budded cells. Dotted line marks 50% disjunction expected for random segregation. (D) Micrographs showing the TRP1 locus (GFP) and total chromosome DNA (DAPI). For cohesion assays and nondisjunction assays, data were derived from two independent experiments, and error bars are SD. For each data point in each experiment 100–400 cells were scored.
Sister chromatid cohesion is required for segregation when spindle assembly is delayed
A likely mechanism for cohesin-independent cohesion arises from some unusual features of budding yeast. During early S phase, a bipolar spindle forms and sister kinetochores achieve stable bipolar attachments concurrent with DNA replication (Pringle et al., 1997; McCarroll and Fangman, 1988; Kitamura et al., 2007). In essence, unreplicated DNA surrounding the duplicated centromeres acts as a surrogate for cohesion. This alternative pathway should be rendered nonfunctional by delaying spindle assembly until after DNA replication. If so, inducing such a delay in cohesion-defective cells should result in random segregation since they rely on this putative alternative pathway.
To test whether this is the case, WT and mcd1-1 cells marked at a CEN4-proximal (TRP1) were synchronously released from G1 phase at 35.5°C in the presence of nocodazole. Treatment with nocodazole blocks spindle assembly and induces mid-M-phase arrest. As expected, most WT cells arrested in mid M phase had one GFP signal, whereas mcd1-1 cells had two GFP signals, indicating the presence or absence of cohesion, respectively (Figure 6A). Cells were then washed free of nocodazole and grown in YPD to allow cells to assemble spindles and enter anaphase (see Materials and Methods). Disjunction of sister chromatids was scored in large-budded cells as described. Most WT cells had one GFP signal in each cell body and equal-sized DNA masses indicative of proper disjunction (Figure 6, B and C). In contrast, mcd1-1 cells had two GFP signals randomly distributed (∼50% disjunction) in cells (Figure 6B). Moreover, DNA masses were unequal and located throughout the two cell bodies (Figure 6C). Thus, delaying spindle formation resulted in random segregation in mcd1-1 cells. There results are consistent with our hypothesis that S phase–coupled spindle assembly provides an alternative pathway enabling cohesion-defective cells to segregate sister chromatids.
FIGURE 6:
Random segregation of sister chromatids in mcd1-1 cells released from nocodazole arrest. WT (VG3460-2A) and mcd1-1 mutant (VG3456-2C) cells arrested in mid M phase 35.5°C using nocodazole and then released from arrest (23°C). Chromosome IV monitored at a CEN4-proximal locus (TRP1). (A) Cohesion at mid-M-phase arrest. Percentage of cells with two GFP signals plotted for G1 and mid-M-phase cells. (B, C) Large-budded cells assayed 2 h after release from nocodazole. (B) Disjunction of chromosome IV sisters. Dotted line marks 50% disjunction expected for random segregation. (C) Micrographs showing chromosomal DNA (DAPI) and CEN-proximal locus of chromosome IV sisters (GFP). For cohesion and nondisjunction assays, 200–400 cells were scored for each data point.
Wpl1p and Smc3p acetylation regulate condensation independent of cohesion
Cohesin-independent segregation provides the means for cells with dramatic defects in establishment to segregate their sister chromatids. However, it remained unclear why smc3-RQ and eco1Δ wpl1Δ mutant cells are viable, whereas K113Q cells are inviable, given that they exhibit the same large cohesion defect. This differential viability suggests that the smc3-RQ and eco1Δ wpl1Δ strains are able to execute an essential cohesin-mediated function that K113Q cells cannot. We speculated that condensation could be that essential function, as cohesin is required for mitotic chromosome condensation in budding yeast (Guacci et al., 1997; Lavoie et al., 2004).
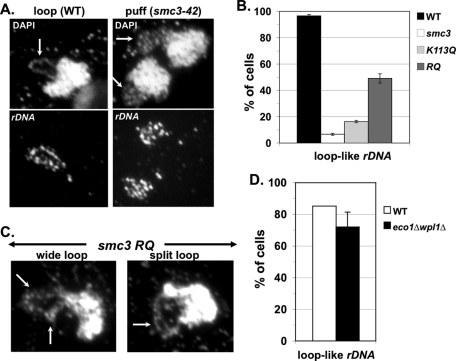
To assess this possibility, we first asked whether the RQ allele differs from the K113Q allele in its ability to mediate condensation. Mutant smc3-42 cells alone or bearing an additional SMC3 WT, K113Q, or RQ allele were synchronously released from G1 phase at 35.5°C and arrested in mid M phase using nocodazole. Cells were fixed and subjected fluorescence in situ hybridization (FISH) to assay condensation of the rDNA locus (see Materials and Methods), which is commonly used to monitor chromosome condensation in budding yeast (Guacci et al., 1993, 1994, 1997; Hartman et al., 2000; Lavoie et al., 2002, 2004). Mid M phase–arrested WT cells had a single rDNA loop adjacent to the bulk DNA (Figures 7, A and B), indicative of sister chromatids that are condensed and have cohesion (Guacci et al., 1993, 1994, 1997). In contrast, in smc3-42 cells, the rDNA formed an amorphous cap, indicative of a failure to property condense (Figures 7, A and B), similar to that previously seen in the mcd1-1 cohesin mutant (Guacci et al., 1997). In the K113Q allele–bearing cells, the rDNA formed an amorphous signal similar to the case of the smc3-42 alone (Figure 7B). In contrast, in the RQ-bearing cells, the rDNA formed loop-like structures in ∼50% of cells, indicative of restored condensation (Figure 7, A and B). Of interest, the rDNA loops formed in the RQ cells were not a single tight loop as in the WT, but rather appeared as either split loops or wide loops (Figure 7C). Split loops likely represented condensed rDNA with precociously separated sisters. Wide loops were likely partially condensed, but we could not assess the status of cohesion.
FIGURE 7:
smc3-RQ allele–bearing smc3-42 cells and eco1Δ wpl1Δ cells promote chromosome condensation at the rDNA locus. (A–C) Haploid strains from Figure 2A, smc3-42 parent alone (smc3) or containing a second SMC3 allele, WT, K113Q, or RQ, were synchronously arrested in mid M phase at 35.5°C using nocodazole as described in Figure 2A. The rDNA resides in the nucleolus adjacent to the bulk chromosomal DNA. (A) Micrographs of WT and smc3-42 cells subjected to FISH. Bulk chromosomal DNA (DAPI) and rDNA detected using FISH (rDNA). Arrows indicate rDNA in DAPI-stained images. (B) Quantitation of rDNA condensation by scoring loop-like rDNA structure. (C) Micrographs of RQ allele in smc3-42 cells subjected to FISH. DAPI-stained bulk chromosomal DNA shown with arrows indicating rDNA. (D) WT and eco1Δ wpl1Δ cells from Figure 3A subjected to FISH as described in A and quantified as described in B. Data were derived from two independent experiments, and error bars are SD. For condensation assays, 50–150 nuclei were scored for each data point in each experiment.
Budding yeast chromosome condensation also requires Eco1p activity, since eco1-ts/ctf7-ts mutant is defective in condensation (Skibbens et al., 1999). Therefore we examined whether deletion of WPL1 could enable condensation without ECO1 inactivity. For this purpose we examined rDNA condensation in eco1Δ wpl1Δ cells arrested in mid M phase. The number of eco1Δ wpl1Δ mutant cells that exhibited loop-like rDNA was close to that of WT cells (Figure 7D). Like the RQ allele–bearing cells, eco1Δ wpl1Δ mutant cells contained rDNA as either split loops or wide loops rather than the single tight loop seen in WT cells. Thus the ability to condense the rDNA in the smc3-RQ cells and the eco1Δ wpl1Δ double mutant, but not the K113Q cells, correlates with their ability to restore viability. These results are consistent with the idea that condensation is the essential cohesin function executed in the smc3-RQ allele–bearing cells and eco1Δ wpl1Δ cells. Moreover, it supports the idea that Eco1p-mediated acetylation and Wpl1p play roles in regulating chromosome condensation.
DISCUSSION
Here we study the roles of Smc3p acetylation, the Eco1p acetyltransferase, Wpl1p, and cohesin in cohesion establishment, as well as in the greater context of sister chromatid disjunction, condensation, and cell viability. We show that eco1Δ wpl1Δ cells establish cohesion very poorly, if at all. This failure to establish cohesion contradicts the prevailing Wpl1p-centric model, which was based upon the incorrect assumption that the ability of a wpl1Δ to restore viability to an eco1Δ mutant serves as a surrogate marker for cohesion establishment (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009). Our result demonstrates that Eco1p promotes S phase cohesion establishment by a mechanism other than, or in addition to, antagonizing Wpl1p. One additional function for acetylation may be modulating the Smc3p ATPase (Unal et al., 2008). Consistent with this, Smc3p K113 residue is predicted to be in close proximity to the Walker A/B domain (Unal et al., 2008). Genetic analyses support this view, as K113 acetyl-mimics limit the toxicity of Smc3p hydrolysis mutants (Heidinger-Pauli et al., 2010b). Our data do not rule out the possibility that Eco1p-mediated acetylation of other cohesin subunits contributes to establishment.
A second insight into the mechanism responsible for cohesion establishment comes from our observation that smc3 acetyl-mimic alleles fail to establish cohesion when present as the sole functional Smc3p in cells. This failure could reflect a role for temporal regulation in establishment, as Smc3p acetylation normally occurs at the onset of S phase, and Eco1p has been genetically and biochemically linked with the replication fork and fork components (Onn et al., 2008; Skibbens, 2011). However, in both human and yeast cells, the cohesion defect of the acetyl-mimic can be partially overcome by the presence of a second copy of wild-type Smc3p (Unal et al., 2008; Zhang et al., 2008). It is important that, in these cells, Smc3p is unable to undergo temporal acetylation because of abrogation of Eco1p activity. These results suggest that limiting the amount of Smc3p acetylation is as important as, if not more important than, temporal regulation. Indeed, only a subset of Smc3p is acetylated in human and yeast cells, consistent with a regulation to limit acetylation levels (Zhang et al., 2008). Furthermore, HOS1 has been identified as an Smc3p deacetylase in budding yeast, and hos1Δ mutant cells have a slightly increased pool of acetylated Smc3p along with minor defects in cohesion (Beckouet et al., 2010; Borges et al., 2010; Xiong et al., 2010). Thus the ratio of acetylated to nonacetylated Smc3p likely is an important contributor to cohesion establishment whose mechanism needs to be elucidated.
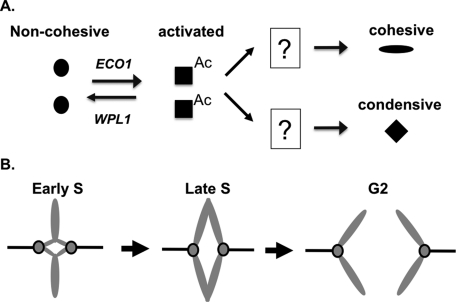
Studies of cohesin regulators such as Eco1p acetylation and Wpl1p have focused on their roles in sister chromatid cohesion. Yet it is known that cohesin and the cohesin regulators Eco1p and Pds5p are required for proper mitotic chromosome condensation in budding yeast (Guacci et al., 1997; Hartman et al., 2000; Lavoie et al., 2002; Skibbens et al., 1999). Here we show that although smc3-RQ, eco1Δ wpl1Δ, and smc3-K113Q strains are equally defective for cohesion, only the smc3-RQ and eco1Δ wpl1Δ strains are competent for condensation of the rDNA locus and are viable. This suggests that Eco1p antagonizes Wpl1p to allow condensation. Smc3p acetylation/removal of Wpl1p inhibition may represent a bifurcation step in which cohesin's fate subsequently diverges, followed by differential regulation of cohesin's cohesive and condensive functions (Figure 8A). The difference in the ability of the RQ and K113Q alleles to promote condensation suggests that, like cohesion establishment, the timing or amount of acetylation may be important for condensation.
FIGURE 8:
(A) Model for regulation of cohesin's roles in cohesion and condensation. Cohesin in noncohesive form (circle). Eco1p acetylates Smc3p to remove Wpl1p inhibition to form activated cohesin (square). Activated cohesin can be used for either cohesion (oval) or condensation (diamond), depending on the activity of unidentified downstream regulators. (B) Model for cohesin-independent segregation. In early S phase, sister kinetochores form bipolar spindle attachments (left). By mid S phase, replicated regions precociously separate, but sister kinetochores retain their bipolar attachments due to unreplicated regions (middle). When replication is completed, poleward movements ensue, enabling sister segregation to opposite poles (right).
The correlation between viability and ability to condense rDNA is consistent with condensation being the essential function executed in the smc3-RQ and eco1Δ wpl1Δ strains. However, Smc3p acetylation could also regulate other, noncohesive functions of cohesin. In fact the smc3-RQ and K113Q strains are sensitive to the DNA-damaging agent camptothecin, suggesting this acetylation may inhibit repair (this study). Budding yeast cohesin has also been shown to play a role in transcriptional regulation and to promote intramolecular loops at pericentric heterochromatin that are believed to help promote segregation (Yeh et al., 2008; Skibbens et al., 2010). We did not assess these functions and so cannot rule out that they contribute to the restoration of viability.
Might acetylation of cohesin regulate condensation in other organisms? Cohesin has been shown to be important in meiotic chromosome condensation in budding and fission yeasts, as well as in mice (Ding et al., 2006; Revenkova et al., 2004; Jin et al., 2009). A recent study using Xenopus egg extracts indicated that cohesin plays a role in condensation when chromosomes were converted into a more mitotic structure by altering the ratio of condensins I and II (Shintomi and Hirano, 2011). Together these results suggest that cohesin plays a conserved role in templating chromosome structure to enable proper mitotic and meiotic condensation in eukaryotes. Therefore it will be interesting to examine the role of cohesin acetylation and Wpl1p in condensation in other organisms. However, metazoans also contain additional proteins that modulate chromosome structure (condensin II complex and sororin) that are not found in budding yeast (Ono et al., 2003; Rankin et al., 2005). These additional proteins may contribute to or alter how acetylation or Wpl1p regulates cohesin's role in condensation.
The ability to condense chromosomes in the smc3-RQ and eco1Δ wpl1Δ strains cannot explain their viability with gross defects in cohesion establishment, since cohesin is assumed to be required for bipolar attachments and segregation. Contrary to this dogma, we show that these strains, as well as those abrogated for Mcd1p and Smc3p function, disjoin sister chromatids reasonably well. This cohesin-independent disjunction likely stems from the unusual biology of budding yeast, in which sister kinetochores and the spindle are assembled early in S phase. The unreplicated DNA around the sister kinetochores acts a surrogate for cohesion, enabling bipolar attachments (Figure 8B). As replication is completed, the sister chromatids precociously separate, yet segregate properly. As predicted by this model, delaying spindle assembly until mitosis abrogates cohesin-independent segregation (this study).
Is this cohesin-independent pathway sufficient on its own to promote viability, or is it supplemented by a small amount of residual cohesin-mediated cohesion that persists in our mutants? We cannot rule out the latter possibility. However, it is clear from the inviability and gross chromosome missegregation that result from inhibiting the S phase–coupled pathway that the cohesin-independent mechanism contributes significantly to viability. In fact our analysis likely underestimates the efficacy of disjunction via the cohesin-independent mechanism. The RQ shows disjunction of 85% of chromosome IV by our assay. If this were the true rate of disjunction on all chromosomes, one would not expect any viable cells, so our assay must underestimate disjunction. One possibility is that smaller chromosomes may disjoin better than chromosome IV, which is the second longest yeast chromosome. Because smc3-RQ and eco1Δ wpl1Δ strains only partially restore condensation, larger chromosomes may be more prone to tangling and delayed segregation. Even though we underestimated the fidelity of the cohesin-independent mechanism for disjunction under unperturbed growth, this pathway is very sensitive to microtubule inhibitors (this study; Sutani et al., 2009). Therefore cohesin-independent segregation is unlikely to be robust enough to enable viability when confronted with environmental stresses found commonly in nature, such as spindle damage or low temperatures.
Is cohesin-independent segregation unique to budding yeast? This replication-coupled mechanism is reminiscent of bacteria, where early-replicating regions segregate before replication is complete (Draper and Gober, 2002). Although spindle assembly in S phase is an unusual feature of budding yeast, the principle of cohesin-independent segregation is worth considering in other eukaryotic organisms as well. For example, DNA catenation can link sisters without cohesin (Vagnarelli et al., 2004), and so, in organisms in which catenation persists into mitosis, such as fission yeast (Uemura et al., 1987), cohesin-independent segregation may exist and influence chromosome segregation and viability.
Mutations in cohesin and its regulators have been implicated in cancer and in developmental diseases, making the elucidation of cohesin regulation of medical relevance (Wang et al., 2004; Dorsett, 2007). For developmental diseases, the severity of the disease state does not correlate with the level of defective cohesion, implicating cohesin's noncohesive functions and illustrating the importance of elucidating their regulation (Dorsett and Krantz, 2009). However, the essential role of cohesin in sister chromatid cohesion rendered it difficult to assess the importance of cohesin's noncohesive functions to cell viability. Our discovery that budding yeast cells are viable without efficient cohesion establishment provides a platform with which to genetically dissect noncohesive functions of cohesin.
MATERIALS AND METHODS
Yeast strains, media, and reagents
Yeast strains used in this study are A364A background, and their genotypes are listed in Supplemental Figure S4. Synthetic complete minimal and YPD media were prepared as described (Guacci et al., 1997). For plates used to assess drug sensitivity, Benomyl (a gift from DuPont, Wilmington, DE) made as a 10 mg/ml stock (in DMSO) was added to a final concentration of 10 μg/ml in media cooled to 55°C. Camptothecin (Sigma-Aldrich, St. Louis, MO) was made as a 4 mg/ml stock (in DMSO) and added to final concentration of 15 μg/ml YPD media containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4.
Plasmids
For SMC3 allele integrations, plasmid pEU40 (SMC3 URA3) and its derivatives containing acetyl-mimics were linearized by SmaI digestion and transformed into relevant yeast strains to integrate at the ura3-52 locus. These plasmids were previously described (Unal et al., 2008), except for the K112R,K113Q (RQ) derivative, which we made using site-directed mutagenesis via the Stratagene QuikChange Kit. The RQ mutation was confirmed by sequencing the entire open reading frame, as well as the promoter region.
For plasmid shuffle, plasmid pEU42 (SMC3 URA3 CEN) and the test pEU41 test plasmid (SMC3 CEN LEU2) and its acetyl-mimic alleles were previously described, except for the RQ allele, which was made and confirmed as described for pEU40 RQ (Unal et al., 2008). Plasmid pRS315 (CEN LEU2) was the empty vector test plasmid.
Dilution plating assays
Cells were grown to saturation in YPD media at 23°C (or 30°C when listed), diluted to OD600 1.0 using YPD, and then plated in 10-fold serial dilutions. Cells were incubated on plates at relevant temperatures as described. For plasmid shuffle, cells were grown at 30°C in SC-LEU media to saturation to allow loss of plasmid pEU42 (SMC3 CEN URA3) but retain test plasmids and then diluted to OD600 1.0 using SC-LEU and plated in fivefold serial dilutions.
Synchronous release from G1 arrest
G1 release into mid-M-phase (nocodazole) arrest.
Cells were grown to mid log phase at 25°C in YPD media, and then α factor (Sigma-Aldrich) was added to 10−8 M and cells incubated 3 h more to induce arrest in G1 phase. These cells were incubated at 35.5°C for 1 h to inactivate temperature-sensitive mutations while still arrested in G1 phase. Cells were washed three times in YPD (35.5°C) containing 0.1 mg/ml Pronase E (Sigma-Aldrich) and once in YPD (35.5°C), resuspended in YPD (35.5°C) containing nocodazole (Sigma-Aldrich) at 15 μg/ml final, and then incubated at 35.5°C for 2 h to arrest in mid M phase. For cultures grown at 30°C, arrest with α factor took 2.5 h, and then cells were washed directly and released without temperature shift as described, except using 30°C YPD.
G1 release to allow one complete cell cycle.
Cells were grown and arrested in G1 phase using at 25°C, shifted to 35.5°C, and washed as described. Cells were resuspended into YPD (35.5°C) and then, after cells had budded (60–70 min), α factor was added to 10−8 M and incubation continued at 35.5°C.
Microscopy
Images were acquired with an Axioplan2 microscope (100× objective, numerical aperture 1.40; Zeiss Thornwood, NY) equipped with a Quantix charge-coupled device camera (Photometrics, Tucson, AZ).
Monitoring cohesion using LacO-GFP assay
CEN-distal cohesion is monitored by integrating LacO repeats at LYS4, located 470 kb from CEN4. CEN-proximal cohesion is monitored by integrating LacO at TRP1, located 10 kb from CEN4. Cells were fixed and processed to allow the number of GFP signals in each cell to be scored as previously described (Unal et al., 2008). The percentage of cells with two GFP signals was determined by dividing the number of cells with two GFP signals by the sum of cells with one GFP signal and with two GFP signals and then multiplying by 100. To image bulk chromosomal DNA, fixed cells were incubated for 5 min in 1% Triton X-100 and then resuspended in buffer containing DAPI at 50 μg/ml final concentration.
Scoring of sister chromatid disjunction
Large-budded (telophase) cells.
We scored only large-budded cells with two GFP signals. The percentage at which the chromosome IV sister chromatids disjoined was given by (cells with one GFP signal in each bud/cells with one GFP signal in each bud plus cells with two GFP signals in one bud and zero GFP signals in the other bud) × 100.
Unbudded (G1 phase) cells.
After cell division, disjunction will generate two cells with one GFP spot each, whereas nondisjunction should generate one cell with two GFP spots and one cell with no GFP spots. However, the prolonged arrest in G1 phase at 35.5°C degraded the LacIGFP fusion so that even in WT cells, 30–40% exhibit no GFP signal. To correct for this class, we simply doubled the number of cells with two GFP spots. Therefore the percentage of unbudded cells showing disjunction was given by (cells with one GFP signal/cells with one GFP signal plus 2× the cells with two GFP signals) × 100.
Fluorescence in situ hybridization
FISH was performed as previously described, except the proteinase K concentration was reduced to 10 μg/ml (Guacci et al., 1997). The rDNA probe directly nick translating plasmid pVG303, which is pUC19 containing a 4.6-kb BglII rDNA fragment from p362 bearing the 5′ half of the rDNA repeat, was previously described (Guacci et al., 1994).
Flow cytometry analysis
This was performed as previously described.
Supplementary Material
Acknowledgments
We thank Jill Heidinger-Pauli, Itay Onn, Fred Tan, Elcin Unal, and Lamia Wahba for their critical reading of the manuscript and for helpful comments, as well as all other member of the Koshland lab for their technical support. This work was funded by the Howard Hughes Medical Institute and National Institutes of Health Grant GM092813.
Abbreviations used:
- BEN
benomyl
- CEN
centromere
- CPT
camptothecin
- DAPI
4′,6-di-amidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- FOA
5-fluoroorotic acid
- GFP
green fluorescent protein
- rDNA
ribosomol DNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0696) on December 21, 2011.
REFERENCES
- Aguilar C, Davidson C, Dix M, Stead K, Zheng K, Hartman T, Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. doi: 10.4161/cc.4.9.1997. [DOI] [PubMed] [Google Scholar]
- Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–688. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper GC, Gober JW. Bacterial chromosome segregation. Annu Rev Microbiol. 2002;56:567–97. doi: 10.1146/annurev.micro.56.012302.160729. [DOI] [PubMed] [Google Scholar]
- Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol Cell Biol. 2011;31:1771–1786. doi: 10.1128/MCB.01284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Yamamoto A, Strunnikov A, Kingsbury J, Hogan E, Meluh P, Koshland D. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harb Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010a;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010b;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Jin H, Guacci V, Yu HG. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–725. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;5:2528–2536. doi: 10.4161/cc.5.21.3405. [DOI] [PubMed] [Google Scholar]
- Onn I, Guacci V, Koshland DE. The zinc finger of Eco1 enhances its acetyltransferase activity during sister chromatid cohesion. Nucleic Acids Res. 2009;37:6126–6134. doi: 10.1093/nar/gkp656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Broach JR, Jones EW. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rowland BD, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV. Sticking a fork in cohesin—it's not done yet. Trends Genet. 2011;27:499–506. doi: 10.1016/j.tig.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Marzillier J, Eastman L. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle. 2010;9:1601–1606. doi: 10.4161/cc.9.8.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 2004;5:167–171. doi: 10.1038/sj.embor.7400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–218. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- Xiong B, Lu S, Gerton JL. Hos1 is a lysine deacetylase for the smc3 subunit of cohesin. Curr Biol. 2010;20:1660–1655. doi: 10.1016/j.cub.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.