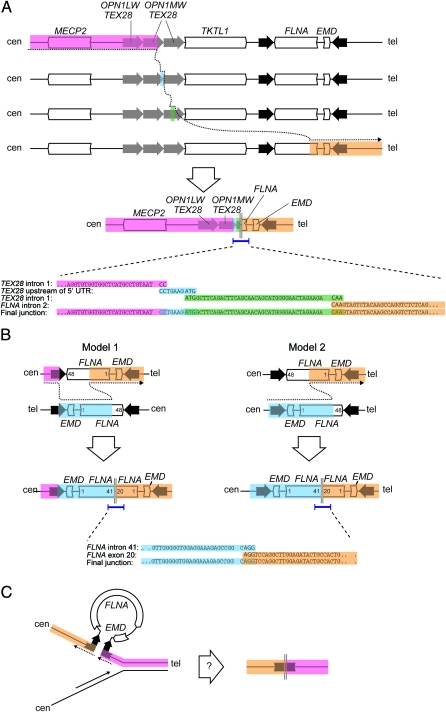
Figure 4. Copy number changes at the FLNA locus in 3 families with periventricular nodular heterotopia implicate distinct molecular mechanisms.
(A) Breakpoint sequencing supports a mechanism for family 1 rearrangement involving replication fork stalling and template switching/microhomology-mediated break-induced replication with template switches occurring between the second OPN1-TEX28 tandem repeat, the junction between the first and second OPN1-TEX28 tandem repeats, the third OPN1-TEX28 repeat, and FLNA intron 2. The result is loss of 1 OPN1-TEX28 tandem repeat and a deletion of all but the 5′ end of FLNA. (B) Breakpoint sequencing in family 2 supports 2 alternative models. In model 1, nonallelic homologous recombination between mispaired inverted repeats is followed by nonhomologous end joining (NHEJ) between 2 inverted copies of FLNA on opposite chromatids, leading to deletion of the 3′ end of FLNA and duplication of the 5′ end of FLNA and EMD. Model 2 presupposes a heterozygous background for the benign FLNA-EMD inversion, in which case a single NHEJ event between FLNA intron 41 on one chromatid and FLNA exon 20 on the second chromatid is sufficient to generate the observed rearrangement. (C) A speculative, replication-based model for FLNA rearrangement in family 3, in which the inverted repeats flanking FLNA and EMD form a single-strand secondary structure that results in contiguous deletion of both genes and the inner half of both repeats.