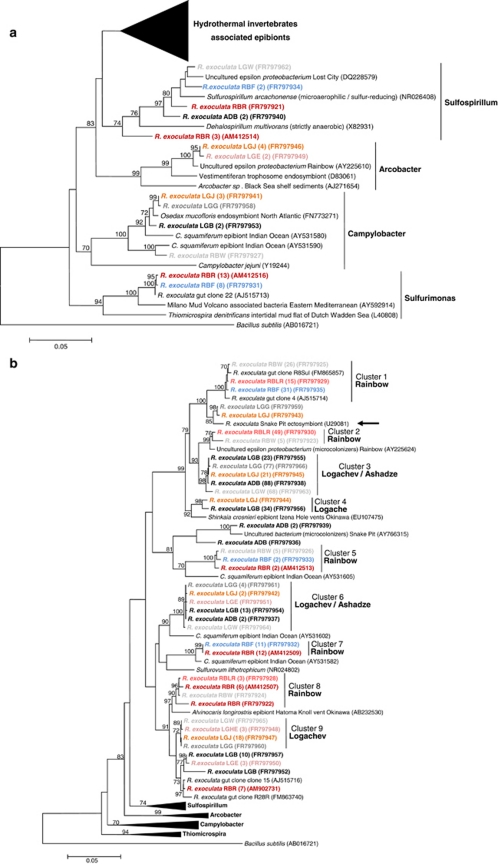
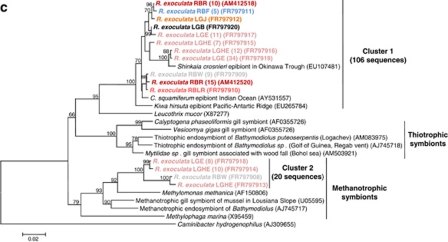
Figure 3.
16S rRNA phylogeny of the Epsilonproteobacteria (a, b; calculated on 817 bp) and Gammaproteobacteria (c; calculated on 804 bp) associated with the R. exoculata gill chamber. Robustness was tested using 500 bootstraps re-sampling of the tree calculated by using the Neighbor-Joining algorithm, using the Kimura two-parameter correction matrix (only bootstrap values over 70 are shown). Sequences names have been resumed as follows: AD, LG or RB for Ashadze, Logachev or Rainbow specimens, respectively, and E, HE, J, W, G, B, LR, R and F for Eggs, Hatched Eggs, Juvenile, White moult, Gray moult, Black moult, Light Red moult, Red moult and Fluid, respectively, and finally the numbers in parentheses refer to the number assigned to each individual. Our clones are shown in color. (a) Global tree representing the R. exoculata epsilon symbiont and their close relatives. (b) Secondary tree showing the ‘Hydrothermal invertebrate-associated epibionts' (Marine Group-1) (see Figure 3a). The black arrow indicates the first R. exoculata epibiont sequence discovered in the Snake Pit site. (c) 16S rRNA phylogeny of the Gammaproteobacteria associated with the R. exoculata gill chamber.