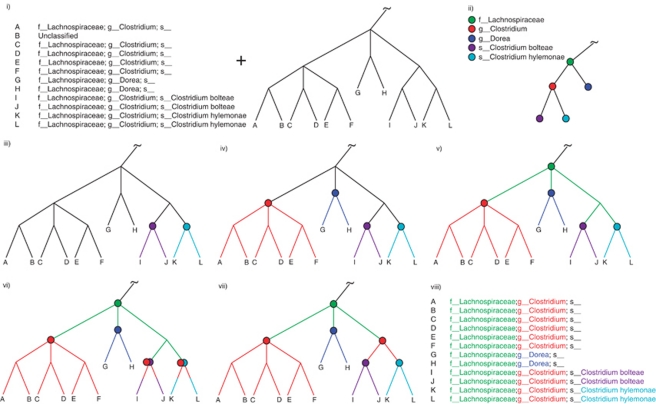
Figure 1.
Overview of the tax2tree workflow. (i) The inputs to tax2tree; a taxonomy file that matches known taxonomy strings to identifiers that are associated with tips of (that is, sequences within) a phylogenetic tree. To simplify the diagram, only the family, genus and species are used, although the full algorithm uses all phylogenetic ranks. (ii) The input taxonomy represented as a tree and a taxon name legend for the figure. (iii, iv) Nodes chosen by the F-measure procedure at each rank; (iii) species, (iv) genus and (v) family. In this example, the genus Clostridium is polyphyletic, and the F-measure procedure picked the ‘best' internal node for the name (uniting tips A–F). However, as unique names at a given rank can only be placed once on the tree, this leaves tips I–L without a genus name placed on an interior node. (vi) The backfilling procedure detects that tips I–L have an incomplete taxonomic path (species to family) and (vi) prepends the missing genus name (obtained from the input taxonomy) to the lower rank because this step of the procedure examines only ancestors but not siblings. (vii) The common name promotion step identifies internal nodes in which all of the nearest named descendants share a common name. In this example, the node that is the lowest common ancestor for tips I–L has immediate descendants that all share the same genus name, Clostridium. This name can be safely promoted to the lowest common ancestor (interior node) uniting tips I–L. (viii) The resulting taxonomy. Note that the sequence identified as B was unclassified in the donor taxonomy but is now classified as f__Lachnospiraceae; g__Clostridium; s__.