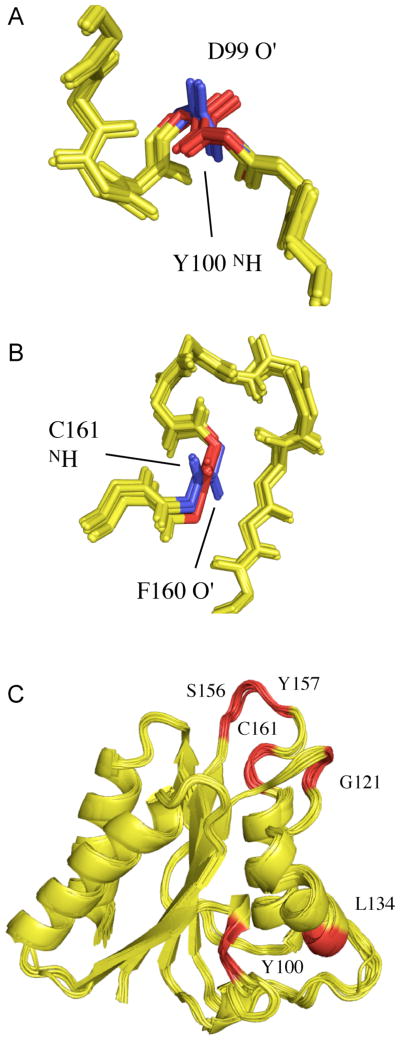
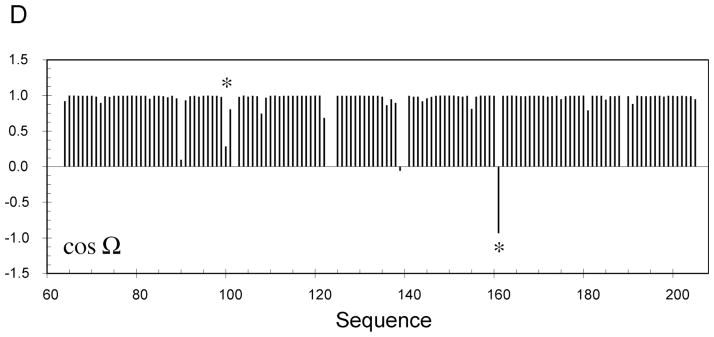
Figure 7.
RDC-consistent orientations of peptide plane units produced from RDC-restrained molecular dynamics calculations using experimental distance restraints from Sulfite Reductase Flavodoxin-like Domain and simulated RDC data from a single alignment tensor. (A) The 20 structures in the lowest experimental energy ensemble sample two RDC-consistent orientations of peptide unit D99/Y100 that are in agreement with the distance constraints. The blue orientation is the correct orientation. (B) The 20 structures in the lowest experimental energy ensemble sample two RDC-consistent orientations of peptide unit F160/C161 that are in agreement with the distance constraints. The blue orientation again shows the correct orientation. (C) Sites of Sulfite Reductase Flavodoxin-like Domain (in red) where two RDC-consistent orientations are observed. (D) Display of cos(Ω) for one of the structures shown in Panels A–C, where Ω is the angle between the peptide planes in the structure of interest and the target, or known structure (see text). The position of peptide planes 99–100 and 160–161 are shown with an asterisk.