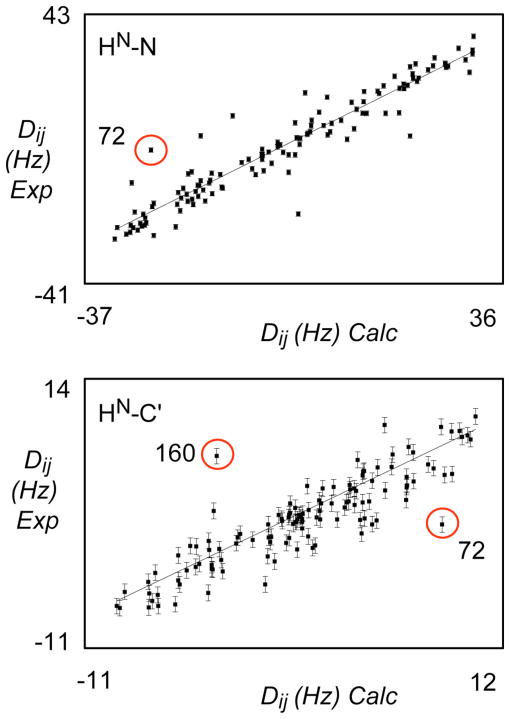
Figure 8.
(A) Best-fit correlation of experimental N-HN RDCs from alignment medium B when fit to optimal alignment tensors determined using structures refined using experimental N-HN, C′-Cα and C′-HN RDCs data from alignment medium A. Correlation from a typical structure is shown from the ensemble best-fitting the active data set (alignment medium A, bacteriophage). A circled outlier corresponds to one of the sites populating two RDC-consistent orientations with respect to these data (see Figure 9).
(B) Best-fit correlation of experimental C′-HN RDCs from alignment medium A when fit to optimal alignment tensors determined using structures refined using experimental N-HN, C′-Cα and C′-HN RDCs data from alignment medium B. Correlation from a typical structure is shown from the ensemble best-fitting the active data set (alignment medium B, alcohol-based alignment). Circled outliers corresponds to sites populating two RDC-consistent orientations with respect to these data (see Figure 9).