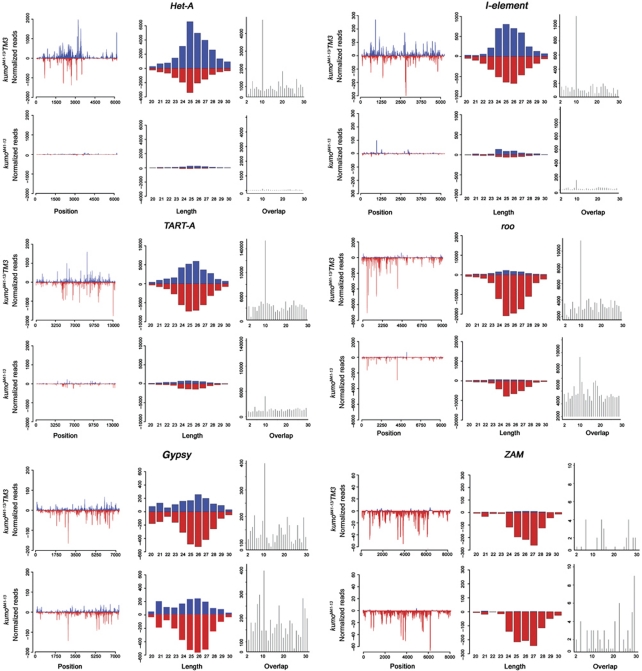
Figure 5.
Reduction of piRNA mapping to transposons in kumo mutant ovaries. Mapping of piRNAs to the sense (blue) and antisense (red) strand plotted over the consensus sequence of various transposon families (left panel); length histogram (right) of all matching RNAs from 20 to 30 nt to sense (blue) and antisense (red) strands; and distribution of overlapping piRNAs (right panel) for 1–30 nt. Loss of kumo function depletes piRNA mapping to the sense and antisense strands of Het-A, I-element and TART-A. Concomitantly, piRNA pairs overlapping with 10 nt of those transposons were almost lost in kumo mutant ovaries. A milder reduction in the amount of piRNAs matching to the antisense strand and having a 10-nt overlap was observed for roo. However, for the transposons targeted by somatic piRNAs, a modest reduction in the number of antisense piRNAs mapping to gypsy and no significant difference in piRNAs levels mapping to ZAM were observed in the kumo mutant.