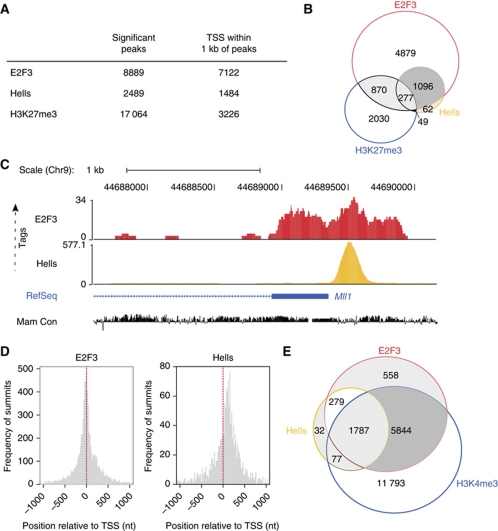
Figure 6.
ChIP-sequencing reveals that E2f3 and Hells share many common target genes. (A) Results of the ChIP-sequencing (ChIP-Seq) for WT MEFs. Genome-wide counts of significant ChIP-Seq peaks for E2f3, Hells and H3K27me3. The number of transcriptional start sites (TSS) within 1 kb of peaks is also given. (B) Venn diagram depicting the contingency between the number of TSS within 1 kb of E2f3, Hells or H3K27me3 ChIP-Seq peaks in WT MEFs. (C) Tag profiles of E2f3 and Hells in WT MEFs in a screenshot from the UCSC genome browser displaying number of ChIP-Seq tags mapping to the Mll1 genomic context. Tag profiles were created by extending each read to a length of 100 bases, and normalizing to 10 million mapped reads. RefSeq transcripts and mammalian sequence conservation (Mam Con) is also shown. (D) Positional preference of the ChIP-Seq peaks for E2f3 and Hells in WT MEFs. Positions are given in nucleotides (nt), and negative values correspond to locations upstream of the TSS. Counts are binned in 20 nucleotide windows. Red-dashed lines indicate the positions of the TSS relative to the peaks. (E) Venn diagram depicting the overlap of E2f3 and Hells-bound promoters (RefSeq) with promoters that are enriched for the trimethylated histone H3 on Lysine 4 (H3K4me3) in WT MEFs.