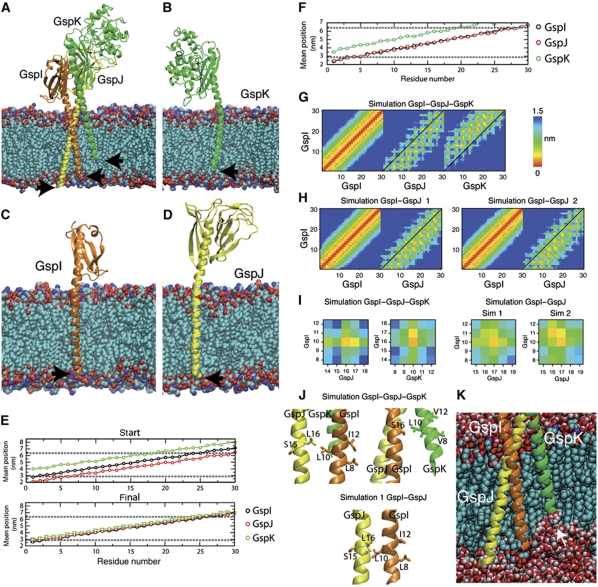
Figure 5.
Molecular dynamics (MD) of full-length GspI, GspJ and GspK in a model membrane. (A) Initial state of the trimeric complex of GspI, GspJ and GspK after adding POPE lipids with arrows indicating the N-termini. (B–D) Snapshot of the final structure of GspI, GspJ or GspK from MD simulations of each protein alone. (E, F) Position of each residue (mean+s.d.) along the bilayer normal in the starting structure and last 5 ns in GspI, GspJ and GspK alone simulations (E) or the GspI–GspJ–GspK complex simulation (F). Dashed lines show the lipid glycerol backbone atoms position. (G, H) Minimum distance between residues in the TM segments of GspI and itself, GspJ or GspK for the GspI–GspJ–GspK complex simulation (G) and two GspI–GspJ dimer simulations (H). (I) As in (G, H) but for residues targeted for crosslinking experiments. (J) Final position of GspI L10, GspJ L16 and GspK L10 after MD simulations. (K) Snapshot of GspI–GspJ–GspK MD simulation showing the membrane deformation induced by GspK.