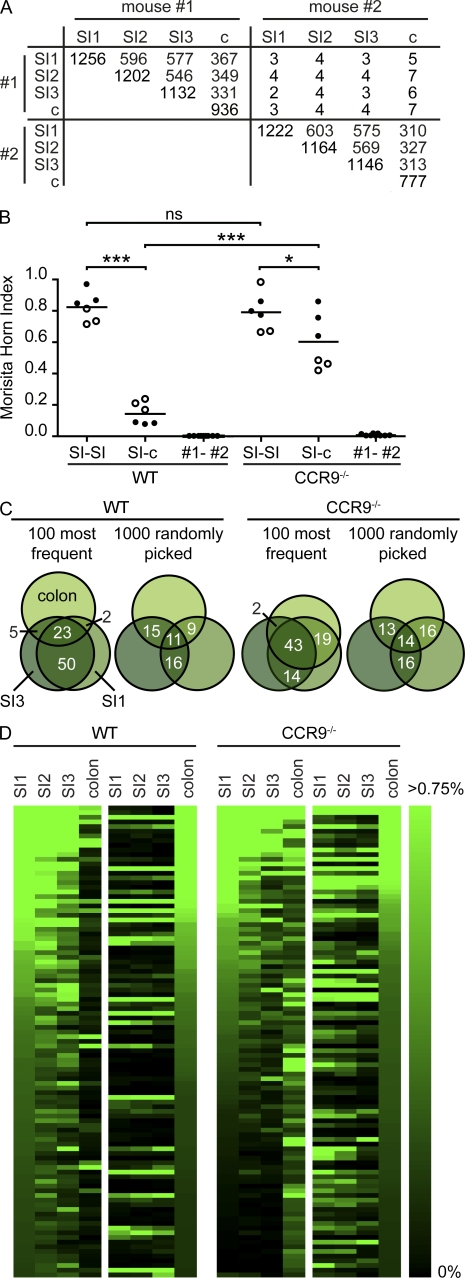
Figure 3.
Identical CDR3 sequences seed different fragments of the SI. (A) Nucleotide CDR3 sequences were compared between proximal jejunum (SI1), distal jejunum (SI2), ileum (SI3), and colon (c) of 13-wk-old C57BL/6 mice, and the number of identical sequences was enumerated. Data are based on 10,000 sequences per sample, and the number of different CDR3 sequences in each sample appears in the diagonal of the table. (B) MHIs were calculated comparing CDR3 sequence pools obtained from various gut fragments in WT and CCR9−/− mice (WT, n = 2; CCR9−/−, n = 2). Individual mice are indicated by open and closed symbols, and comparisons calculated are SI1 to SI2, SI1 to SI3, and SI2 to SI3 to describe similarity of SI fragments and SI1 to c, SI2 to c, and SI3 to c for comparison of SI- and large intestine–derived sequence pools. Comparison of CDR3 sequence pools between different mice yields MHIs close to 0 (first and second columns). Horizontal lines indicate the mean. *, P < 0.05; ***, P < 0.001. (C) Venn diagrams illustrate the overlap of CDR3 sequence pools from SI1, SI3, and colon in one WT and one CCR9−/− mouse. Numbers indicate overlap in percentages. (D) 10,000 CDR3 sequences from SI1, SI2, SI3, and colon were sorted according to their frequency in SI1 (WT, left) or colon (WT, right). Because overlap of sequence sets is driven by expanded sequences, we limited our analysis to the 100 most frequent sequences and visualized their frequency (in percentages of all sequences) by color codes. Note that upon sorting according to frequency in the proximal jejunum, more sequences with similarly high frequency could be found in CCR9−/− mice than in WT mice.