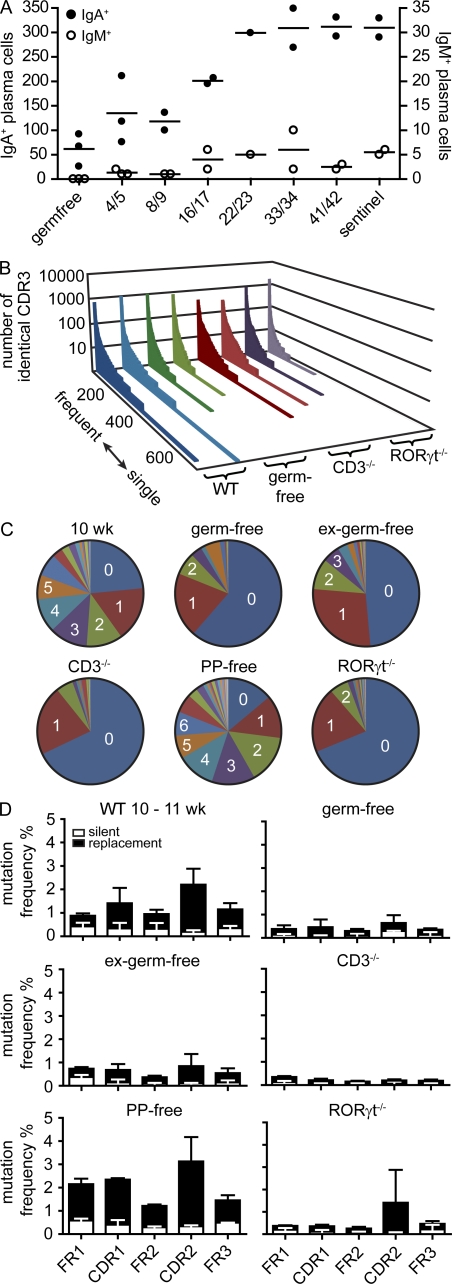
Figure 7.
SHM requires T cells, microbiota, and RORγt but not PPs. (A) Germ-free mice were colonized by cohousing with sentinels raised under SPF conditions for the indicated number of days. Symbols indicate the total number of IgA+ and IgM+ cells in individual mice observed by histology counting six fields of view from at least three different sections per mouse covering different fragments of the SI. Horizontal lines indicate the mean. (B) In a set of 5,000 sequences, all different CDR3 sequences were listed along the x axis from frequent to single sequences, and the number of sequence reads for each different CDR3 sequence was displayed on the y axis. Each slice represents an independently performed experiment as indicated. (C) Diagrams depict the number of SMs observed in representative 10-wk-old WT (see Fig. 5), germ-free, ex–germ-free, CD3−/−, PP-free, and RORγt−/− mice. (D) Bars depict mean + SD SM frequencies in FR1, FR2, FR3, CDR1, and CDR2 observed in the following number of animals: 10–11-wk-old WT mice (n = 6; identical to data in Fig. 5 B and shown to improved readability), germ-free mice (9 wk; n = 2), ex–germ-free mice (11 wk; n = 2), CD3−/− mice (10 wk; n = 3), mice lacking PPs (12 wk; n = 2), and RORγt−/− mice (20 wk; n = 2). For IgA sequences from PP-free mice, RNA was isolated out of sorted IgA+CD138+ intestinal plasma cells. For each mouse, a minimum of 10,000 sequences were analyzed.