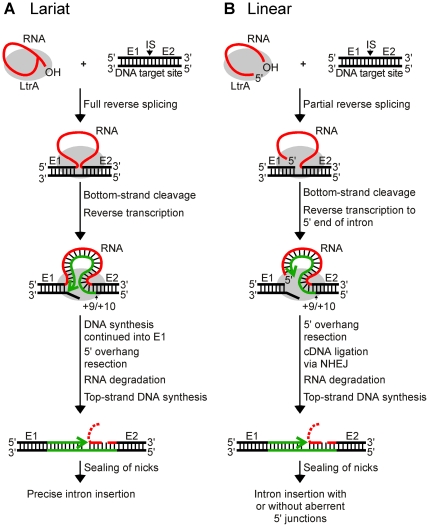
Figure 1. Models for retrohoming of Ll.LtrB group II intron lariat and linear RNAs.
(A) Retrohoming of lariat RNA. RNPs containing lariat RNA recognize the DNA target site (ligated E1–E2 sequence) and carry out both steps of reverse splicing, resulting in insertion of the intron RNA between E1 and E2. The IEP uses its En domain to cleave the bottom strand between positions +9 and +10 of E2, and then uses the 3′ end of the cleaved DNA strand as a primer for reverse transcription of the inserted intron RNA. The resulting full-length intron cDNA is extended directly into E1 by continued DNA synthesis. Retrohoming is completed by a process that includes removal of the 5′ overhang on the bottom strand, degradation or displacement of the intron RNA template strand, top-strand DNA synthesis by a host DNA polymerase, and sealing of nicks by a host DNA ligase [21]. (B) Retrohoming of linear RNA. RNPs containing linear intron RNA recognize the DNA target site and carry out the first step of reverse splicing, resulting in ligation of the 3′ end of the intron RNA to the 5′ end of E2. The IEP then uses its En domain to cleave the bottom strand between positions +9 and +10, generating a primer for reverse transcription of the intron RNA, as in lariat RNA retrohoming. However, because the 5′ end of the linear intron RNA is unattached, the resulting cDNA cannot be extended directly into E1 and is instead linked to the 5′ exon DNA by an error-prone process that sometimes leads to precise insertion of the intron RNA, but often gives imprecise 5′ junctions due to deletion of E1 sequences, 5′-intron truncations, and/or insertion of extra nucleotide residues at the ligation junction. As for lariat RNA, retrohoming of the linear intron RNA is completed by degradation or displacement of the intron RNA template strand, top-strand DNA synthesis, and sealing of nicks by host enzymes. E1 and E2, 5′ and 3′ exon, respectively; CS, bottom-strand cleavage site; IS, intron-insertion site.