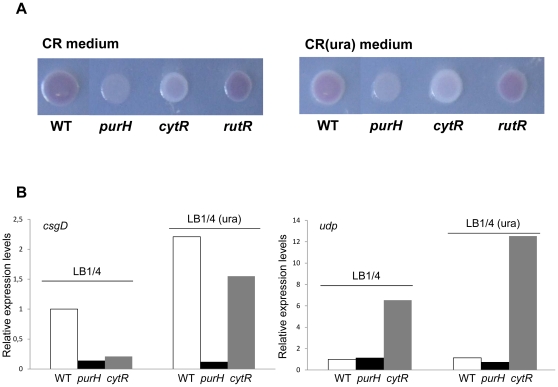
Figure 4. Congo red binding by E. coli strains deficient in pyrimidine sensing (cytR and rutR mutants) and purine biosynthesis (purH mutant).
4A. The MG1655 strain and its isogenic mutants in the purH, cytR and rutR genes were spotted either on CR medium (left panel) or on CR(ura) medium (right panel) and grown for 24 hours at 30°C. Plates were incubated for 48 hours at 4°C to enhance Congo red binding. Determination of transcript levels. 4B. Relative expression of either the csgD gene (left panel) or the udp gene (right panel) was determined by Real-Time PCR on RNA extracted from overnight cultures of MG1655 and of its isogenic purH and cytR mutants. 16S RNA transcript was used as reference gene. ΔCt values between the genes of interest and 16S RNA were set at 1 for MG1655 in LB1/4 medium, and transcript levels in other strains and/or growth conditions are expressed as relative values. Experiments were repeated at least three times, each time in duplicate; standard deviations were always lower than 5%.