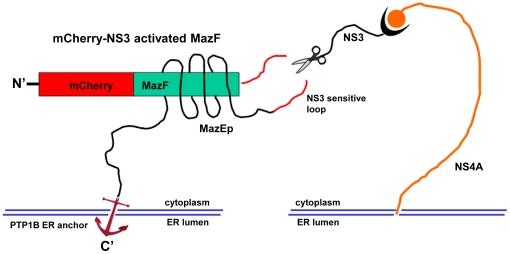
Figure 1. Schematic representation of the construct “mCherry-NS3 activated MazF” and hypothetical mechanism of activation by NS3 protease.
The NS3-activated MazF zymoxin was constructed by fusing 5 elements in the following order (from the N terminus): monomeric red fluorescence protein mCherry, E. coli MazF ribonuclease, HCV P10-P10' NS3 cleavage sequence derived from 2a genotype (strain JFH1) NS5A/B junction, a short inhibitory peptide corresponding to MazE C-terminal 35 amino-acids (which encompass the 23 amino-acids inhibitory peptide (MazEp) that has been described by Li et al. [25]) and the C-terminal ER membrane anchor of the tyrosine phosphatase PTP1B [28]. After being anchored to the ER membrane, the NS3 cleavage site that is located between the ribonuclease and the inhibitory peptide in the “mCherry-NS3 activated MazF” construct (which is active as a dimer but for convenience is illustrated here in its monomeric form) is cleaved by the HCV- NS3 protease which is also localized to the cytoplasmic side of the ER membrane. The toxic ribonuclease, no longer covalently tethered to its ER-anchored inhibitor, is now free to diffuse to the cytoplasm (which lacks the antidote) and exert its destructive activity.