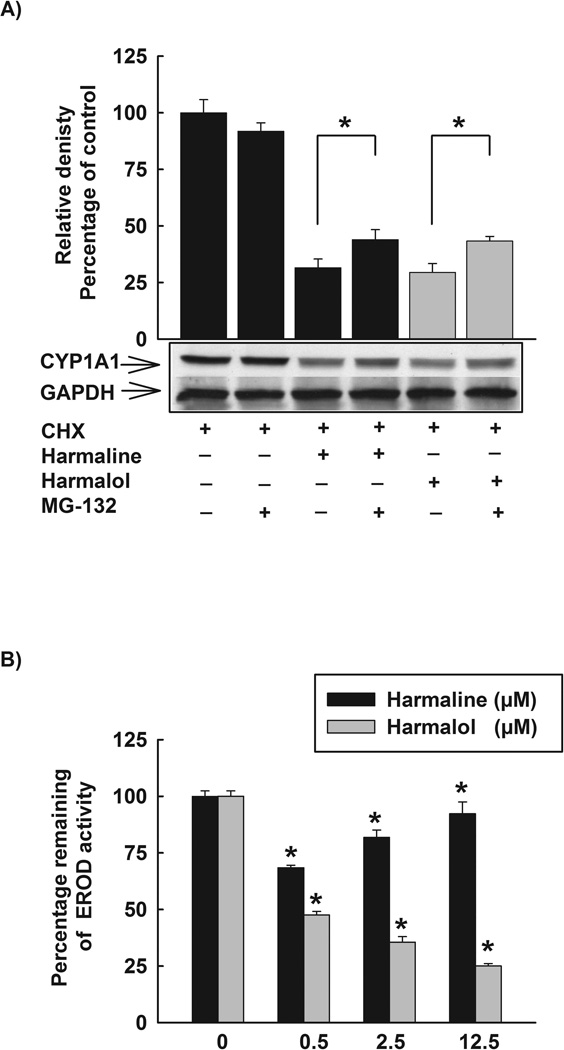
Figure 10. Posttranslational modifications of CYP1A1 by harmaline and harmalol.
(A) Effect of proteasomal inhibitor, MG-132, on the reduced CYP1A1 protein stability by harmaline and harmalol. HepG2 cells were treated with TCDD (1 nM) for 24 h. Thereafter, cells were washed three times with PBS and incubated with fresh media containing CHX (10 µg/mL, the protein translation inhibitor), CHX (10 µg/mL) and harmaline (2.5 µM) or CHX (10 µg/mL) and harmalol (2.5 µM) in the absence and presence of the proteasomal inhibitor, MG-132 (0.5 µM). After 6 h incubation, total protein was extracted and CYP1A1 protein was detected using Western blot analysis. The intensities of CYP1A1 protein bands were normalized to GAPDH signals, which were used as loading controls. Values represent mean of relative densities and expressed as percentage of control (CHX and DMSO treated cells) ± S.E.M. One of three representative experiments is shown. (*) P < 0.05 compared with the relevant treatment. (B) The direct inhibitory effects of harmaline and harmalol on CYP1A1 enzyme. HepG2 cells were pre-treated with TCDD (1nM) for 24 h, thereafter media were removed, washed three times with PBS, and increasing concentrations of harmaline and harmalol (0.5–12.5 µM) in assay buffer (Tris (0.05 M), NaCl (0.1 M), pH 7.8) were added for 15 min prior to the addition of 7ER (2 µM final concentration) for the EROD measurement. Results are expressed as percentage of remaining EROD activity (mean ± S.E.M, n = 8). (*) P < 0.05 compared with control.