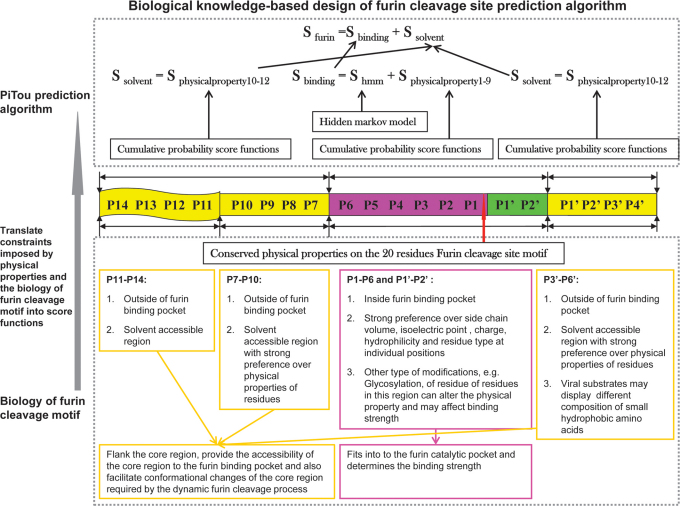
Figure 1.
The design of PiTou algorithm: the PiTou score function 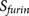 is biological knowledge-based and it comprises of a machine
learning-based hidden Markov model and a set of biological mechanism-based cumulative
probability score functions.
is biological knowledge-based and it comprises of a machine
learning-based hidden Markov model and a set of biological mechanism-based cumulative
probability score functions.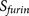 is the sum of two
parts: the core region binding score
is the sum of two
parts: the core region binding score 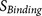 (calculated from eight amino acids at P6–P2′) and the flanking region solvent accessible
score
(calculated from eight amino acids at P6–P2′) and the flanking region solvent accessible
score 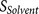 (calculated from eight amino acids at
P7–P14 and four amino acids at P3′–P6′).
(calculated from eight amino acids at
P7–P14 and four amino acids at P3′–P6′). 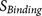 and
and 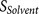 incorporate the physical properties of the
20-residue furin cleavage motif and the binding of substrates to the furin catalytic
domain. The analysis of the 20-residue furin cleavage motif is described in previous
publication8.
incorporate the physical properties of the
20-residue furin cleavage motif and the binding of substrates to the furin catalytic
domain. The analysis of the 20-residue furin cleavage motif is described in previous
publication8.