Figure 2. Determination of optimal binding sequence for p150Sal2.
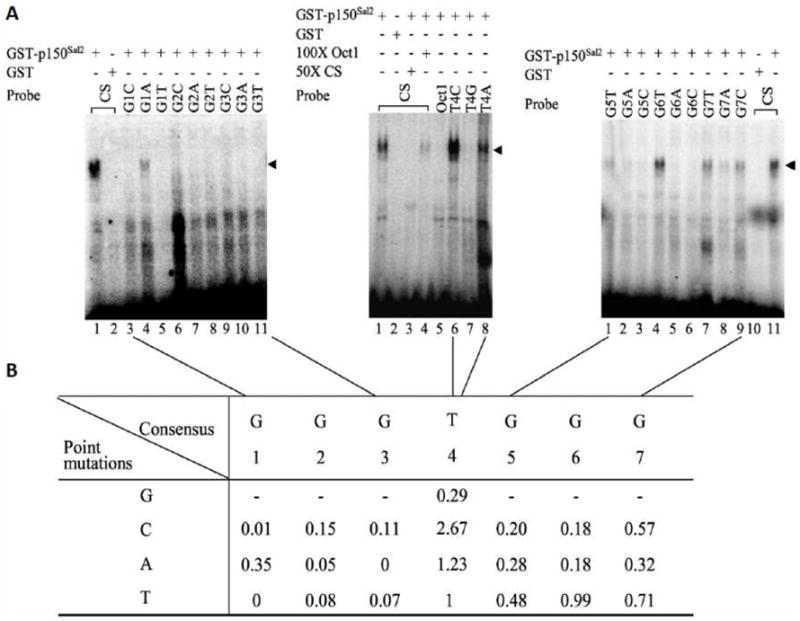
A - EMSA was performed to test the binding efficiency of GST-p150Sal2 towards [α-32P]dCTP-labeled consensus sequence (CS) from SELEX and single base substitutions at each position (left panel, lanes 3-11; middle panel, lanes 6-8; right panel, lanes 1-9). Equal amount (100ng) of GST-p150Sal2 or GST vector was used in the binding reaction. Competition was performed using 50 or 100 fold excess of unlabeled oligonucleotides as specific or non-specific competitor (middle panel, lanes 3 and 4, respectively). B - Summary of binding efficiencies of GST-p150Sal2 to oligonucleotides. Band densities of oligonuleotide-GST-p150Sal2 complexes were quantified using a PhosphorImager and normalized to the SELEX consensus sequence set as 1. GST-p150Sal2-DNA complexes are indicated (◂).
