Figure 3. The triple zinc finger cluster in p150Sal2 is essential for DNA binding.
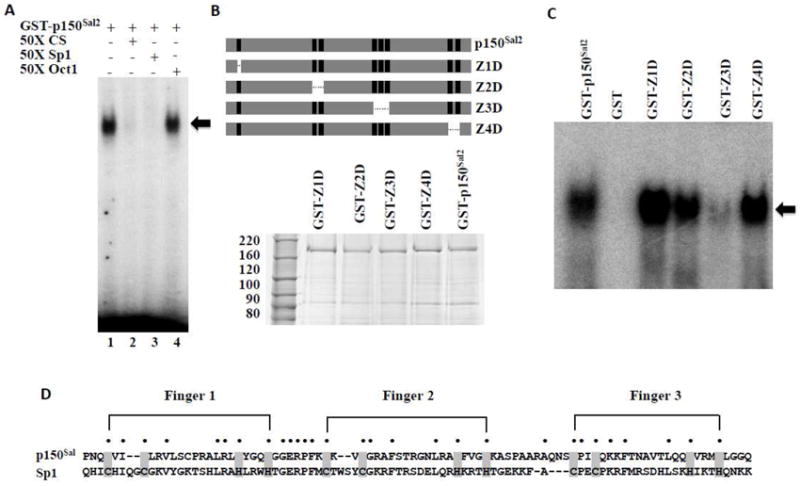
A - Binding of p150Sal2 to an oligonucleotide carrying a confirmed Sp1 binding site (5’-ATTCGATCGGGGCGGGGCGAGC-3’). Competition was conducted by adding 50 fold molar excess of unlabelled oligonucleotides containing the p150Sal2 consensus sequence (5’-GGATCACTGGGTGGGAATCACGCT-3’) (lane 2), Sp1 binding site (lane 3) or Oct 1 binding site (5’-TGTCGAATGCAAATCACTAGAA-3’) (lane 4) in the reaction. DNA-GST-p150Sal2 complexes are indicated (◂). B - Top: Schematic of p150Sal2 illustrating zinc finger deletion mutants. Bottom: Purified GST-p150Sal2 and zinc finger deletion mutants. C - Equal amounts of purified GST-p150Sal2 or mutant proteins were incubated with labelled Sp1 oligonucleotide and complexes analyzed by EMSA (◂). D - Alignment of the triple zinc finger motif in p150Sal2 and Sp1 illustrating similarities of spacing of the C2H2 fingers and linking sequences.
