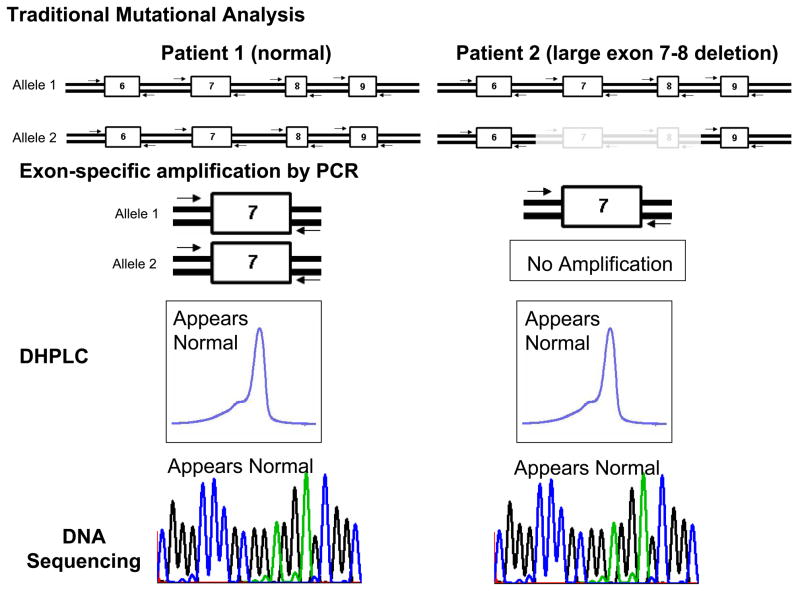
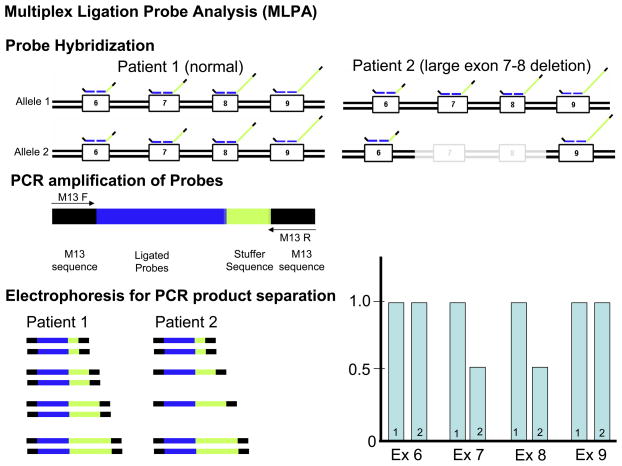
Figure 1. Traditional Mutational Analysis by DHPLC versus MLPA Detection for Large Insertion or Deletion Mutations.
Depicted is A) a traditional mutational analysis by DHPLC where exon specific PCR amplification of both the maternal (1) and paternal (2) alleles is achieved using specific primers designed to anneal to the exon flanking intronic sequences. In the case of normal patient (patient 1), PCR products representing both alleles will be produced. However, in patient 2 who hosts a large deletion, PCR primers will only anneal to the normal strand. In either patient, the resulting DHPLC and subsequent sequence will only be representative of the normal strand and the allele with the deletion will go unnoticed. B) Illustrates how multiplex ligation probe analysis (MLPA) can be used to detect such deletions or insertions by quantifying the copy number of target sequences (exons) as determined by specifically designed probes. Here, specific probes of varying length (as a result of the variable length “stuffer” sequence-shown in yellow) are first hybridized to each patient’s gene sequence. The probes are stripped from the genomic DNA and subsequently serve as template for PCR amplification using primers that specifically anneals to all of the probes allowing them to be simultaneously amplified in a single multiplex PCR reaction. Because of the varying length of the “stuffer” sequence, the amplified probes can be separated according to size by electrophoresis and viewed on a chromatogram. The amount of probe hybridizing to the patient’s genomic DNA determines the copy number for the specific targeted sequence (exons). In this example where exon 7 and 8 are deleted, less exon 7 and 8 specific probes have been hybridized and subsequently fewer copy numbers of template are detected. Conversely, a large insertion would produce more copy numbers of the target sequence(s).