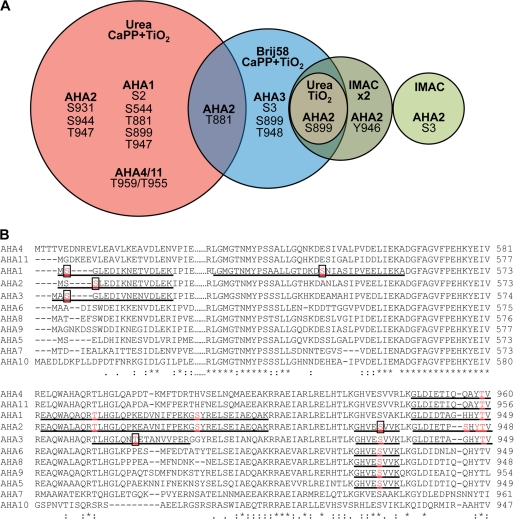
FIGURE 1.
Phosphosites identified in plasma membrane H+-ATPases homologously expressed in A thaliana membranes. A, phosphosites were determined in A. thaliana plasma membrane H+-ATPases using different approaches for phosphopeptide enrichment. Plasma membranes purified from A. thaliana seedlings were exposed for trypsin digestion directly, after treatment with Brij58, or after solubilization by urea/thiourea. IMAC, TiO2, and calcium phosphate precipitation followed by TiO2 (CaPP+TiO2) were used for phosphopeptide enrichment. Phosphorylation sites in the phosphopeptides were validated manually and are unique for the isoform of AHA mentioned in this figure with following exceptions: Site Ser-931 of AHA2 was determined in the peptide GHVEpSVVK, which can derive also from AHA2, AHA3, AHA5, AHA6, AHA8, AHA9, and Thr-959/955 in the peptide GLDIETIQQAYpTV that is shared by AHA4 and AHA11 isoforms. All the phosphopeptides determined are presented in the supplemental material. B, shown is ClustalW analysis of parts of the N terminus (N), P domain (P), and C terminus (C) of the ATPase isoforms of Arabidopsis. The identified phosphopeptides are underlined, and phosphorylated residues printed in bold and red. Novel residues are boxed.