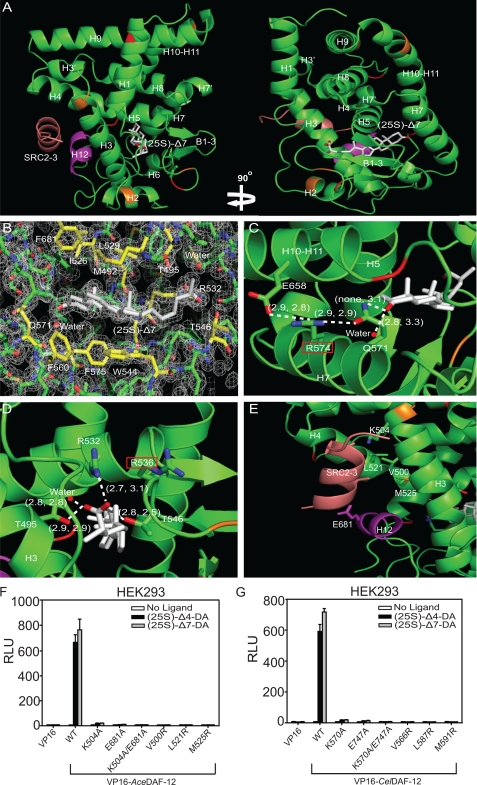
FIGURE 2.
Structure analysis of the AceDAF-12 LBD. A, ribbon model reveals the overall architecture of the AceDAF-12 LBD chain a (green) complexed with (25S)-Δ7-DA (white) and the coactivator peptide SRC2-3 (brown). The AF-2 helix is highlighted in magenta. Mutated cysteines are in red and mutated lysines in orange. B, electron density map of the AceDAF-12 a chain ligand binding pocket bound to (25S)-Δ7-DA. The surrounding amino acids are highlighted in yellow. See supplemental Fig. S2 for the electron density map of complex b. C, H-bonding of AceDAF-12 amino acids and water involved in binding the C3-keto group of (25S)-Δ7-DA. Arg-574 (red box) serves as the H-bonding network hub. D, H-bonding of AceDAF-12 amino acids and water to the C27-carboxyl group of (25S)-Δ7-DA. Arg-536 (red box) can compensate Arg-532 for interacting with the ligand. H-bonds are illustrated by white dashed lines with bond lengths noted in Å. The first number in parentheses indicates the bond length in complex a and the second number indicates bond length in complex b. E, amino acids involved in binding the coactivator peptide SRC2-3 (salmon) are labeled. F and G, mutation of the labeled amino acids (E) in AceDAF-12 (F) or corresponding amino acids in CelDAF-12 (G) disrupts the binding to SRC2-3 LXXLL coactivator motifs in mammalian two-hybrid assays performed in HEK293 cells. 1 μm (25S)-DA is used. RLU is relative light unit that is normalized with β-galactosidase activity as transfection control.