Background: SfmD is a hypothetical protein thought to be a hydroxylase in saframycin A biosynthesis.
Results: SfmD binds heme and hydroxylates tyrosine analogs using H2O2 as oxidant.
Conclusion: SfmD is a heme peroxidase that catalyzes hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine using H2O2 as oxidant.
Significance: This study reveals a novel type of heme peroxidase and has significance for the biosynthesis of analogues of saframycin A.
Keywords: Biosynthesis, Enzyme Catalysis, Heme Oxygenase, Hydroxylase, Peroxidase, Saframycin, SfmD, Heme-containing Peroxidase, Hydrogen Peroxide, Hydroxylation
Abstract
Saframycin A (SFM-A) is a potent antitumor antibiotic that belongs to the tetrahydroisoquinoline family. Biosynthetic studies have revealed that its unique pentacyclic core structure is derived from alanine, glycine, and non-proteinogenic amino acid 3-hydroxy-5-methyl-O-methyltyrosine (3-OH-5-Me-OMe-Tyr). SfmD, a hypothetical protein in the biosynthetic pathway of SFM-A, was hypothesized to be responsible for the generation of the 3-hydroxy group of 3-OH-5-Me-OMe-Tyr based on previously heterologous expression results. We now report the in vitro characterization of SfmD as a novel heme-containing peroxidase that catalyzes the hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine using hydrogen peroxide as the oxidant. In addition, we elucidated the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr by kinetic studies of SfmD in combination with biochemical assays of SfmM2, a methyltransferase within the same pathway. Furthermore, SacD, a counterpart of SfmD involved in safracin B biosynthesis, was also characterized as a heme-containing peroxidase, suggesting that SfmD-like heme-containing peroxidases may be commonly involved in the biosynthesis of SFM-A and its analogs. Finally, we found that the conserved motif HXXXC is crucial for heme binding using comparative UV-Vis and Magnetic Circular Dichroism (MCD) spectra studies of SfmD wild-type and mutants. Together, these findings expand the category of heme-containing peroxidases and set the stage for further mechanistic studies. In addition, this study has critical implications for delineating the biosynthetic pathway of other related tetrahydroisoquinoline family members.
Introduction
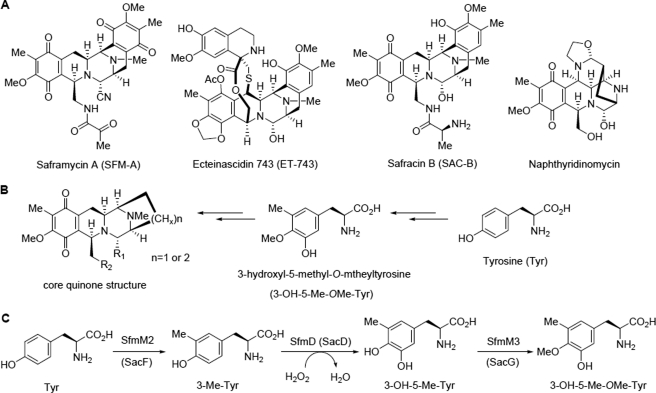
Tetrahydroisoquinoline alkaloids are a growing class of antibiotics possessing a characteristic tetrahydroisoquinoline structure (1). Members of this family display a range of antitumor, antimicrobial, and other biological activities depending on their structures. Saframycin A (SFM-A,3 Fig. 1A), isolated from Streptomyces lavendulae NRRL 11002, is a representative member of this family with potent antitumor activity that is derived from iminium ion via a carbinolamine moiety (2). The most well-known member of this family of compounds, ecteinascidin 743 (ET-743, Fig. 1A), which is a highly potent analog of SFM-A, has been approved as an anticancer drug for the treatment of advanced soft tissue sarcoma (1, 3). It shares a central pentacyclic core with SFM-A, with the exception of the oxidation state of their terminal rings and an additional macrolactone bridge found in ET-743. Moreover, most of the tetrahydroisoquinoline family members, such as saframycins, safracins, naphthyridinomycins, and ecteinascidins, all possess the same core quinone structure (Fig. 1B), which implies that there may be some common precursors involved in their biosynthesis (1).
FIGURE 1.
Structures of representative tetrahydroisoquinoline family members with their common features and the proposed biosynthetic pathway of 3-OH-5Me-OMe-Tyr. A, structures of the representative members of tetrahydroisoquinoline family natural products. B, core quinone structure of these members and its deduced biosynthetic precursors. C, proposed biosynthetic pathway of 3-OH-5-Me-OMe-Tyr involved in SFM-A or SAC-B biosynthesis.
A series of biosynthetic studies have indicated that the quinone moiety is derived from the non-proteinogenic amino acid precursor 3-hydroxy-5-methyl-O-methyltyrosine (3-OH-5-Me-OMe-Tyr), which originates from tyrosine (Tyr) (4–9). Recently, a three-gene cassette, sfmD/sfmM2/sfmM3 from the biosynthetic gene cluster of SFM-A or sacD/sacF/sacG from the biosynthetic gene cluster of SAC-B, was reported to be responsible for the biosynthesis of 3-OH-5-Me-OMe-Tyr (6–8). The hypothetical proteins, SfmD and its homolog SacD, were deduced to be hydroxylases responsible for the 3-hydroxy group construction based on heterologous expression results (7, 8). However, these proteins have no sequence similarity to the known characterized proteins, and little is known about these enzymes or the reactions that they catalyze.
Generally, several types of hydroxylases that catalyze hydroxylation on the aromatic ring of Tyr or its derivatives have been reported (supplemental Fig. S1). One major type is pteridine dependent, which is represented by Tyr hydroxylase that is involved in the biosynthesis of catecholamine neurotransmitters (10). Another type is tyrosinase, which is copper dependent and involved in the biosynthesis of melanin pigments (11). The third type is two-component, FAD-dependent monooxygenase, such as SgcC, which was reported to catalyze the hydroxylation of β-Tyr tethered to a carrier protein during the biosynthesis of the enediyne antitumor antibiotic C-1027 (12). A common feature of these types of Tyr hydroxylases is that all of them are non-heme-dependent monooxygenases and catalyze the hydroxylation using oxygen as the oxidant.
Distinct from these types of Tyr hydroxylases, we herein report the in vitro biochemical characterization of SfmD as a heme-containing peroxidase that catalyzes the hydroxylation of Tyr and its derivatives using hydrogen peroxide (H2O2) as the oxidant. From kinetic studies of SfmD and enzyme assays of SfmM2, the true substrate of SfmD was identified as 3-methyltyrosine (3-Me-Tyr), and the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr was also elucidated. This type of heme-containing peroxidase was further demonstrated to be a potentially common factor in the biosynthesis of SFM-A and its analogues, which possess the same core quinone structure as SFM-A, as determined by characterization of SacD in vitro. Furthermore, comparative analyses of UV-Vis and Magnetic Circular Dichroism (MCD) spectra of SfmD wild-type and SfmD mutants revealed that the conserved motif HXXXC plays a crucial role in heme binding.
EXPERIMENTAL PROCEDURES
General Methods, Biochemicals, and Chemicals
DNA isolation and manipulation in Escherichia coli were performed according to standard methods (13). PCR amplifications were carried out on an authorized thermal cycler (Eppendorf AG 22331, Hamburg, Germany) using PrimeSTAR HS DNA polymerase (TaKaRa, Japan). Primer synthesis and DNA sequencing were performed at Shanghai Invitrogen Biotech Co., Ltd. (China). The E. coli DH5α cells were purchased from Invitrogen (Carlsbad, CA), and E. coli BL21 (DE3) cells were purchased from Novagen (Madison). Restriction enzymes were purchased from TaKaRa Biotechnology Co., Ltd. (Dalian, China). All chemicals and reagents were purchased from Sigma-Aldrich or GL Biochem (Shanghai) Ltd. unless noted otherwise. Analytical HPLC was carried out on an Agilent 1200 HPLC system with PDA detector. LC-MS analysis was carried out on an Agilent 1200 HPLC instrument connected to LCQ Fleet electrospray ionization (ESI) mass spectrometer (Thermo Fisher Scientific Inc.). The electrospray ionization-mass spectroscopies (ESI-MS) were performed with a Shimadzu LCMS-2010 EV mass spectrometer. NMR data were collected using a Bruker 400 MHz spectrometer. Tyr, 3,4-dihydroxyphenylalanine (Dopa), O-methyltyrosine (OMe-Tyr), and dl-m-Tyr were purchased from Sigma-Aldrich.
Chemical Synthesis of Tyr Derivatives
The 3-methyl-O-methyltyrosine (3-Me-OMe-Tyr) and 3-OH-5-Me-OMe-Tyr were each prepared as described (6, 14, 15). Synthesis of 3-Me-Tyr and 3-hydroxy-5-methyltyrosine (3-OH-5-Me-Tyr) were achieved by following a previously reported method (6, 14, 15). The experimental details and characterization of the intermediates are supplied under supplemental materials. The 1H NMR assignments of 3-Me-Tyr are as follows: 1H NMR (400 MHz, D2O): δ 2.18 (s, 3H), 3.06∼3.25 (m, 2H), 4.21∼ 4.24 (m, 1H), 6.85 (d, 1H, J = 8.0Hz), 7.01 (d, 1H, J = 8.0Hz), 7.08 (s, 1H); MS (ESI): m/z 196.18 ([M+H]+). The 1H NMR assignments of 3-OH-5-Me-Tyr are as follows: 1H NMR (400 MHz, D2O): δ 2.21 (s, 3H), 3.04∼3.23 (m, 2H), 4.24∼4.34 (m, 1H), 6.67 (s, 1H), 6.70 (s, 1H); MS (ESI): m/z 212.0 ([M+H]+).
Cloning, Expression, and Purification of SfmD
The sfmD gene was PCR-amplified from plasmid pTL2027 (8) as a template with the primers listed in supplemental Table S1. The purified PCR product was cloned into the EcoRI/HindIII site of pSP72 to generate pTL2039. When the sequence was verified, a 1.1 kb NdeI/HindIII fragment recovered from pTL2039 was ligated into the same site of pET37b to afford the expression plasmid pTL2040. C-terminal His8-tagged SfmD was overexpressed in E. coli BL21 (DE3). A 3 ml starter culture was grown overnight from a single colony in LB media with 50 μg ml−1 kanamycin at 37 °C with shaking and then used to inoculate 1 liter of LB medium at 37 °C with 50 μg ml−1 kanamycin for 2 h. The cultures were then cooled to 16 °C for 1 h, induced with 0.1 mm IPTG when the A600 reached 0.5, and incubated at 16 °C for an additional 24 h.
All protein purification steps took place at 4 °C. Protein purification buffers contained 500 mm NaCl, 50 mm sodium phosphate adjusted to pH 8.0, 10% glycerol and increasing concentrations of imidazole. Buffers A (lysis), B (wash), and C (elution) contained 10, 50, and 250 mm imidazole, respectively. The cells were harvested by centrifugation (6000 rpm for 5 min at 4 °C) and resuspended in buffer A. The cells were lysed by sonication (10 × 60 s pulsed cycle), and the debris was removed by centrifugation (12,000 rpm for 60 min at 4 °C). Soluble protein was collected and purified on a Ni-NTA column by washing with several volumes of buffer B and eluting with 10 ml of buffer C. The eluant was concentrated and desalted into 50 mm Tris-HCl buffer (pH 8.0), 50 mm NaCl, and 10% glycerol using a PD-10 column (GE Healthcare). The purified proteins were concentrated using an Amicon Ultra-4 (10K, GE Healthcare), stored as 10% glycerol stocks at −80 °C, and utilized without further modifications. Protein purity was assessed by 12% acrylamide SDS-PAGE. Protein concentration was determined by the Bradford method (16) using a BSA calibration curve.
A modificatory procedure to overproduce SfmD in the presence of 5-aminolevulinic acid (ALA) was similar to the aforementioned procedure, with the exception that ALA (1 mm, final concentration) and (NH4)2Fe(SO4)2 (1 mm, final concentration) were added to the culture at the same time as IPTG. The heme content of purified SfmD was determined by the pyridine hemochrome assay as previously described (17).
In Vitro Enzymatic Assays of SfmD
The assay conditions used to test whether SfmD is a heme-containing oxygenase are similar to the assay conditions that reported for heme-containing oxygenases in the presence of oxygen, such as conditions for cytochrome P450 (18), secondary amine oxygenase (19), heme oxygenase (20), prostaglandin H Synthase (21), and tryptophan 2, 3-dioxygenase TioF (22).
The assay conditions used to test SfmD as a heme-containing peroxidase are as follows. Standard assay (at 25 °C) mixtures (50 μl) were composed of 100 mm Tris-HCl, pH 9.0, 1 mm ascorbic acid, 2 mm H2O2, 1 mm 3-Me-Tyr (Tyr, OMe-Tyr, or 3-Me-OMe-Tyr), and 50 μm SfmD. In the H218O2 assays, only H2O2 was replaced by H218O2. The reactions were started by adding H2O2 and terminated by adding 0.5 μl of trichloroacetic acid (TCA). Identical assays with boiled SfmD were carried out as negative controls. Following centrifugation to remove protein, the reactions were analyzed by HPLC or LC-MS using an analytic Inertsil ODS-EP column (5 μm, 4.6 × 250 mm, GL Sciences). The LC conditions were as follows. Solvent A was H2O, and Solvent B was CH3CN; both solvent contained 0.1% TFA (v/v) in the HPLC analysis or 0.1% HCO2H (v/v) in the LC-MS analysis. The column was equilibrated with 100% solvent A and followed by the following linear gradient program: 100% A/0% B, 0–3 min; ramp to 28% B, 3–15 min; ramp to 95% B, 15–17 min; and return to 100% A/0% B, 17–20 min. The flow rate was 1 ml min−1, and elution was monitored at 276 nm.
The optimized reaction conditions for SfmD were determined by varying the assay pH (5.0–9.5). Kinetic studies of the protein were performed by changing the substrates (Tyr or 3-Me-Tyr) concentrations from 25 to 2000 μm under optimized conditions with 4 mm H2O2. Product formation was determined using HPLC. Each data point represents a minimum of three replicate end point assays; kinetic constants were obtained by nonlinear regression analysis using OriginLab OriginPro (OriginLab software, Northampton, MA).
Cloning, Expression, Purification, and Biochemical Characterizations of SacD
The construction procedure for the expression plasmid of sacD was similar to sfmD. Plasmid pTL2031 (8) was used as the PCR template. The resulting expression plasmid pTL2044 is a derivative of pET37b, in which SacD will be overproduced as a C-terminal His8-tagged fusion protein. The expression and purification of SacD were carried out in a manner similar to the modificatory procedure of SfmD. The purified proteins were utilized without further modifications. The biochemical characterizations of SacD were carried out similar to the procedures of SfmD.
Cloning, Expression, and Purification of SfmM2
The construction procedure for the expression plasmid of sfmM2 was similar to sfmD. Plasmid pTL2027 (8) was used as the PCR template. The resulting expression plasmid pTL2042 is a derivative of pET37b. C-terminal His8-tagged SfmM2 was overexpressed in E. coli BL21 (DE3). The expression and purification procedures of recombinant SfmM2 were carried out in a manner similar to that described for SfmD. The purified proteins were utilized without further modifications.
Characterization of the C-methylation Function of SfmM2 in Vitro
Standard reactions (50 μl) consisted of 100 mm Tris-HCl (pH 8.0), 2 mm SAM, 1 mg ml−1 BSA, 1 mm Tyr (OMe-Tyr, 3-Me-OMe-Tyr, or dl-m-Tyr), and 50 μm SfmM2 at 30 °C. The reactions were started by adding SfmM2 and terminated by adding 0.5 μl of TCA. Identical assays with boiled SfmM2 were carried out as negative controls. Following centrifugation to remove protein, the reactions were analyzed by HPLC or LC-MS using an analytic Inertsil ODS-EP column (5 μm, 4.6 × 250 mm, GL Sciences). The LC conditions were as follows. Solvent A was H2O, and Solvent B was CH3CN; both solvent contained 0.1% formic acid (v/v) in the HPLC or LC-MS analysis. The column was equilibrated with 100% solvent A and followed by a linear gradient program: 100% A/0% B, 0–3 min; ramp to 16% B, 3–20 min; ramp to 95% B, 20–23 min; and return to 100% A/0% B, 23–26 min. The flow rate was 1 ml min−1 and elution was monitored at 276 nm.
Construction of SfmD Mutants
The specified mutants of SfmD were performed by site-specific mutagenesis using the QuickChange Muti-Site Directed Mutagenesis Kit (Stratagene) according to the manufacture's introductions. The pTL2039 vector was used as the template, and the primers listed in supplemental Table S2 were used for the specified mutant. Consequently, the mutated versions of sfmD were confirmed by sequencing and then cloned into pET37b using the same strategy to make pTL2040 for native SfmD expression respectively. Protein expression and purification of these mutants were carried out in a manner similar to the modification procedure described for SfmD wild type (WT). The enzyme assays of mutants were also similar to the assays of SfmD WT.
Spectroscopic Techniques
UV-Visible absorption spectra were recorded with a Jasco V530 spectrophotometer at room temperature. MCD spectra were measured in 0.1-cm cuvettes at a magnetic field of 0.75T with the Jasco J810 spectropolarimeter at 4 °C as previously described (23). MCD manipulations were carried out as previously reported (23) using JASCO software. The electron paramagnetic resonance (EPR) spectra of SfmD WT were carried out on a Bruker EMX plus 10/12 spectrometer system (Bruker Co., Ltd., Germany) at the High Magnetic Field Laboratory, Chinese Academy of Sciences, Hefei, China. Samples were prepared in Tris-HCl (100 mm, pH 7.5) buffer to appropriate concentrations for SfmD WT and its mutants.
RESULTS
Bioinformatics Analysis of SfmD
BLAST analysis of the 365-amino acid sequence of SfmD revealed that SfmD had no sequence similarity to any functionally characterized proteins in the NCBI database, and no putative conserved domain was identified. There were only three hypothetical proteins that showed homology to SfmD: SacD, which is involved in safracin biosynthesis (51% similarity), AZL_f00720 from Azospirillum sp. B510 (45% similarity), and KSE_08540 from Kitasatospora setae KM-6054 (43% similarity). Because no useful information was obtained, a structural homology search was performed using the online program HHpred, which revealed that the C terminus of SfmD was structurally homologous to tryptophan 2, 3-dioxygenase (TDO) from Ralstonia metallidurans and indoleamine 2, 3-dioxygenase (IDO) from human. Both of these proteins are heme-containing oxygenases that catalyze the oxidative cleavage of tryptophan to N-formyl kynurenine. However, further investigation was needed to determine whether SfmD is a heme-containing protein.
Enzyme Purification and Cofactor Identification of SfmD
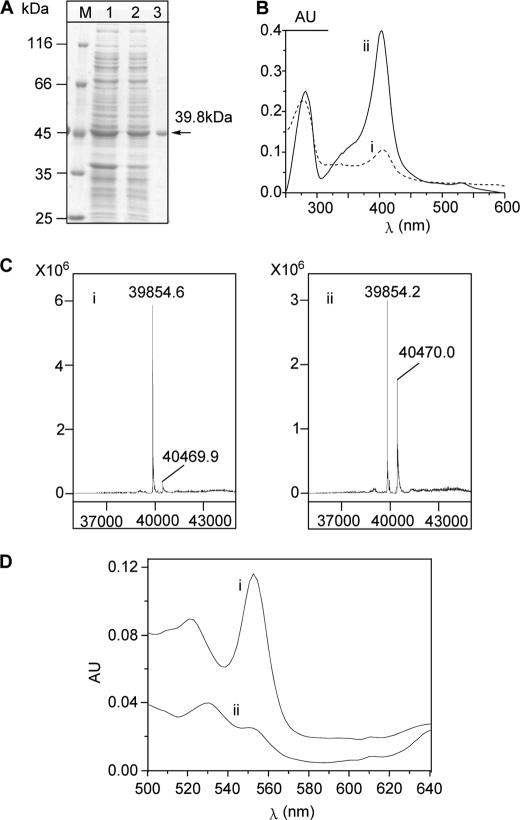
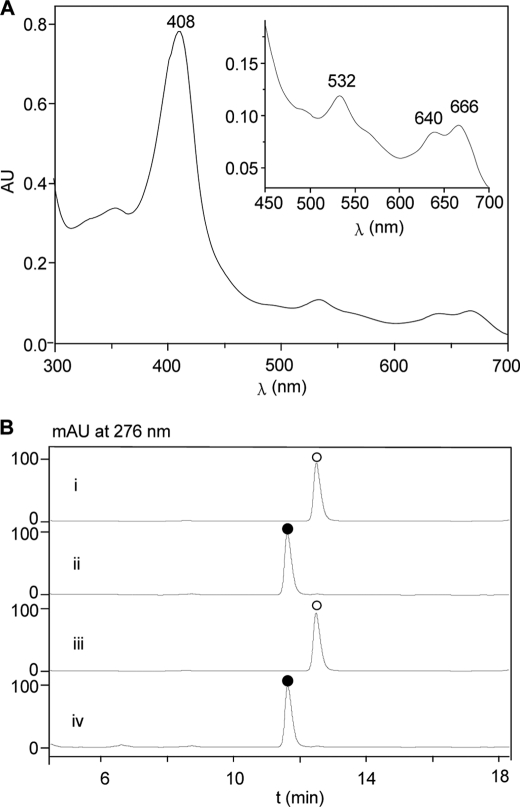
Recombinant SfmD was expressed in E. coli BL21 (DE3) for in vitro characterization. The C-terminal His8-tagged enzyme was purified by Ni-NTA affinity chromatography and migrated as a single band at the predicted size of 39.8 kDa based on SDS-PAGE analysis (Fig. 2A). The purified SfmD was green and exhibited absorbance maxima at 404 nm in UV-Vis spectra (Fig. 2B), which were similar to other heme-containing proteins reported in literature (24, 25). When subjected to LC-MS analysis, the major peak was found to be 39,854.6 Da (Fig. 2C), which is in accordance with the calculated molecular weight (MW) of SfmD (39,854.12 Da) without any cofactor. Meanwhile, another minor peak, 40,469.9 Da, which represents an additional 615.3 Da, was also detected, indicating that parts of the proteins may be co-purified with cofactors. Among the known cofactors, it was close to the calculated MW of heme (616.43 Da). We then overexpressed and purified SfmD again using a modified procedure where 5-aminolevulinic acid (ALA), a biosynthetic precursor of heme, was added to the culture broth at the time of protein expression induction. As expected, not only was the absorption intensity at 404 nm in UV-Vis spectra remarkably increased (Fig. 2B), but the abundance of the peak at 40,470 Da in the mass spectrum was also increased (Fig. 2C). Together, these data indicated that SfmD is a heme-containing protein. To quantify the amount of heme bound to the protein, pyridine hemochrome assays (17) were carried out. The results showed that 5 μm SfmD bound 4.6 ± 0.2 μm heme (mean ± S.D., n = 3) (Fig. 2D), indicating that one SfmD binds one molecular of heme.
FIGURE 2.
Protein purification and cofactor identification of SfmD. A, purification of recombinant C-His8-tagged SfmD as monitored by SDS-PAGE. Lane M, molecular weight marker; lane 1, total proteins after IPTG induction; lane 2, total soluble proteins; lane 3, purified protein. B, UV-Vis spectra of SfmD. SfmD without (i) and with (ii) addition of ALA when the protein was induced to express. C, characterization of SfmD by LC-MS. SfmD without (i) and with (ii) addition of ALA during the protein expression. D, pyridine hemichrome and hemochrome assays of SfmD, the amount of heme bound to SfmD was calculated by following the absorbance change at 553 nm using a difference extinction coefficient of 23.76 mm−1 cm−1 (17). (i) the hemochrome of SfmD; (ii) the hemichrome of SfmD.
Characterization of SfmD as a Heme-containing Peroxidase
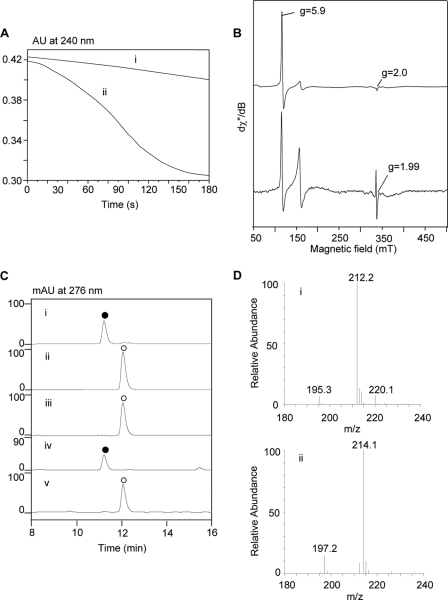
Heme-containing proteins generally fall into two large superfamilies: heme-containing oxygenases, which usually use oxygen as the oxidant, and heme-containing peroxidases, which usually use peroxides as the oxidant (25, 26). Because SfmD showed structural homology to TDO and IDO, we first tested whether SfmD was an oxygen-dependent protein. Various assays of SfmD with the four plausible substrates (Tyr, 3-Me-Tyr, OMe-Tyr, and 3-Me-OMe-Tyr) were carried out using the assay procedures reported in the literature for heme-containing oxygenases in the presence of oxygen (see “Experimental Procedures”). Unfortunately, no corresponding hydroxylation products were detected under these conditions (data not shown). We then examined whether SfmD can use H2O2 as the oxidant. To our surprise, when SfmD was added into the solution of H2O2, a slow dismutation of H2O2 occurred (Fig. 3A), demonstrating that SfmD has very weak catalase activity (the turnover number was only ∼0.6 s−1 calculated from the oxygen electrode experiment, see supplemental information). Additionally, low temperature (10 K) EPR was used to characterize the properties of SfmD. Spectra were recorded before and after the addition of H2O2 to the enzyme in the absence of reducing substrates. The spectrum of SfmD was dominated by the high-spin ferric signals (g⊥ = 5.9 and g‖ = 2.0, Fig. 3B), which was similar to the EPR spectra of the resting peroxidases (27). After the addition of H2O2, the spectra of peroxide-activated SfmD showed a free radical with g ∼1.99, which was in agreement with what was previously assigned to the porphyrin radical in heme peroxidase Compound I (28). Moreover, when we incubated 3-Me-Tyr with SfmD in the presence of H2O2, it was completely converted to a new compound that had an HPLC retention time and MS that were identical to the authentic product 3-OH-5-Me-Tyr (Fig. 3C). To confirm that the oxygen atom that was inserted in the product originated from H2O2, H218O2 assays were conducted with 3-Me-Tyr and resulted in an [M+2] product as expected (Fig. 3D). Therefore, these data provided in vitro validation that SfmD is a heme-containing peroxidase that catalyzes the regioselective hydroxylation of aromatic amino acid using H2O2 as the oxidant.
FIGURE 3.
Characterization of SfmD as a heme-containing peroxidase. A, H2O2 dismutation experiments recorded in UV-Vis spectra at 240 nm. The solution of H2O2 in the absence (i) or presence (ii) of SfmD (25 μm). B, low temperature EPR spectra of SfmD (50 μm). Spectra of SfmD resting state (top) indicating the high-spin ferric signals (g = 2.0 and g = 5.9) and its peroxide-activated form (Compound I) (bottom). C, HPLC analysis of the hydroxylation of 3-Me-Tyr to 3-OH-5-Me-Tyr catalyzed by SfmD: authentic standard of 3-OH-5-Me-Tyr (i) and 3-Me-Tyr (ii), reaction with O2 (iii) and H2O2 (iv) as the oxidant, negative control with boiled enzyme (v); (●), 3-OH-5-Me-Tyr, (ο), 3-Me-Tyr. D, LC-MS analysis of the production of 3-OH-5-Me-Tyr using H2O2 (i) and H218O2 (ii) as the oxidant.
Substrate Specificity and Kinetics of SfmD
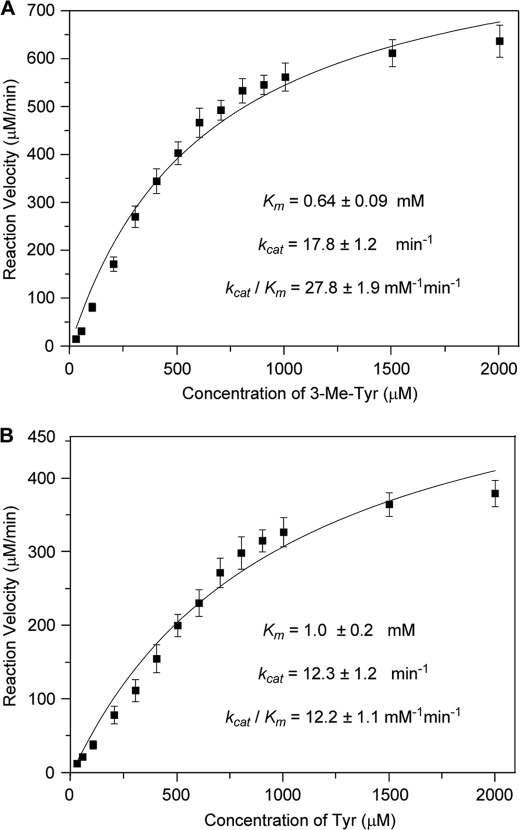
To identify enzyme substrate specificity, the four plausible substrates were assayed. As shown in supplemental Fig. S2, both Tyr and 3-Me-Tyr could be hydroxylated, indicating that SfmD had some substrate flexibility. For kinetic analysis, assay conditions were studied at varying pH values (ranging from 5.0 to 9.5 in 0.5 steps). The optimized condition was found to be at pH 9.0 (supplemental Fig. S3). Under this condition, the kinetic studies toward Tyr and 3-Me-Tyr were carried out in the presence of excess H2O2 (Fig. 4). SfmD showed a conversion with a Km of 0.64 ± 0.09 mm and kcat of 17.8 ± 1.2 min−1 toward 3-Me-Tyr and Km of 1.0 ± 0.2 mm and kcat of 12.3 ± 1.2 min−1 toward Tyr. The higher specificity (kcat/Km, shown in Fig. 4) of SfmD toward 3-Me-Tyr over Tyr indicated that 3-Me-Tyr should be the true substrate of SfmD in vivo.
FIGURE 4.
Kinetic analysis of the hydroxylation reaction catalyzed by SfmD with substrate 3-Me-Tyr (A) or Tyr (B). The reaction mixtures consist of 50 μm protein, excess H2O2 (4 mm) and variable substrate (25 μm-2 mm) under optimized conditions.
Confirming the Biosynthetic Pathway of 3-OH-5-Me-OMe-Tyr
To further investigate the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr, SfmM2, the putative C-methyltransferase, was overexpressed and purified as an N-His8 tagged recombinant protein. The enzyme migrated as a protein with a MW of ∼42 kDa by SDS-PAGE analysis, which was close to the predicted MW of SfmM2 (41.8 kDa) (supplemental Fig. S4A). In vitro biochemical assays using this recombinant protein indicated that only Tyr can be converted to the corresponding methylated product 3-Me-Tyr among the tested substrates (supplemental Fig. S5). Based on the assay results of SfmD and SfmM2, we confirmed the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr as follows (Fig. 1C): Tyr is first methylated to 3-Me-Tyr by SfmM2, and then SfmD catalyzes the hydroxylation of 3-Me-Tyr to 3-OH-5-Me-Tyr. Finally, 3-OH-5-Me-Tyr is transformed to 3-OH-5-Me-OMe-Tyr by the putative O-methyltransferase SfmM3.
Enzymatic Characterizations of SacD
From the biosynthetic pathway of SAC-B, there is a homolog of SfmD that was named as SacD. To test whether the properties or functions of SacD were similar to SfmD, we overexpressed the protein in E. coli and purified it as a C-His8 tagged recombinant protein (supplemental Fig. S4B). The purified SacD showed a maxima absorbance at 404 nm in UV-Vis spectra (Fig. 5A) and a weak ability to dismutate H2O2 (data not shown). In the in vitro assays, SacD also used H2O2 as the oxidant, displayed an optimal activity at pH 9.0 (supplemental Fig. S6), and converted 3-Me-Tyr to the corresponding hydroxylation product, 3-OH-5-Me-Tyr (Fig. 5B). These results indicated that, like SfmD, SacD is also a heme-containing peroxidase that catalyzes the hydroxylation of aromatic amino acids using H2O2 as the oxidant.
FIGURE 5.
UV-Vis spectra of SacD (A) and HPLC analysis of the hydroxylation of 3-Me-Tyr to 3-OH-5-Me-Tyr catalyzed by SacD (B). (i) Authentic standard of 3-Me-Tyr; (ii) authentic standard of 3-OH-5-Me-Tyr; (iii) negative control with boiled enzyme; (iv) reaction mixture; (●), 3-OH-5-Me-Tyr, (○), 3-Me-Tyr.
Determination of the Heme-binding Site of SfmD
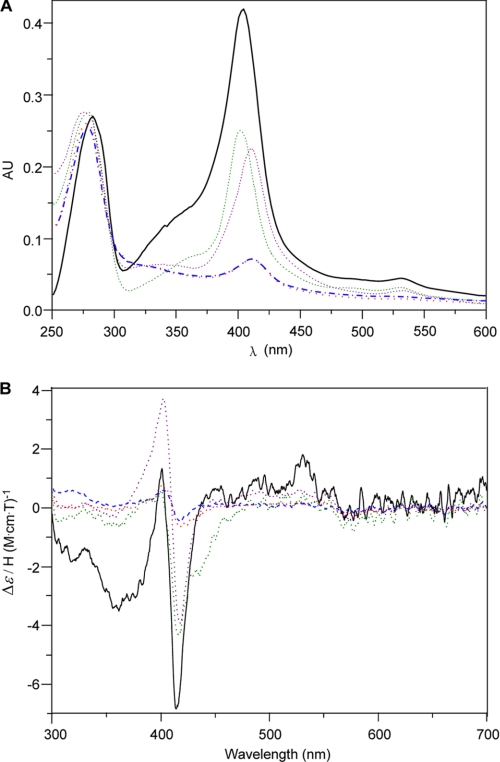
In heme-containing proteins, there are usually one or more conserved motifs that are crucial for heme binding, such as FXXGXXXCXG in P450s (29), CXXCH in c-type cytochromes (30), HPGG in cytochrome b5 (31), CPs in proteins regulated by heme (32), GXXDG in DyP-type peroxidases (24), and others. Sequence alignment of SfmD and its analogous (SacD, AZL_f00720, and KSE_08540) did not identify any of these conserved motifs, but three other conserved motifs were found, including WXXXH, SGXXXXDH, and HXXXC (Fig. 6). To test whether these motifs are important for heme binding, we constructed several mutants of SfmD, such as H191A, H274A, H313A, and C317A, by site-specific mutagenesis, since the residues His and Cys are usually essential for heme binding. These mutants were overexpressed and purified in E. coli using the same conditions as SfmD WT (supplemental Fig. S7). When the UV-Vis spectra were measured, the absorbance intensity was dramatically decreased in SfmD H313A and SfmD C317A compared with SfmD WT, SfmD H191A and SfmD H274A (Fig. 7A), suggesting that the heme content of SfmD H313A and SfmD H317A may be very low. Furthermore, the MCD spectra of SfmD WT and these mutants demonstrated that the signal intensity of SfmD WT, SfmD H191A, and SfmD H274A were comparable and the signal intensity of SfmD H313A and SfmD C317A were very weak (Fig. 7B), indicating that the mutants H313A and C317A had very little heme-Fe binding ability. Together, these data confirmed that the conserved motif HXXXC found in SfmD is crucial for heme binding. In addition, we found that all the four mutants of SfmD lost the ability to hydroxylate 3-Me-Tyr (supplemental Fig. S8), suggesting that the other two conserved motifs also play important roles in the catalytic cycles.
FIGURE 6.
Multiple sequence alignment of SfmD and its analogs. Amino acid sequences were obtained from GenBankTM, including: SfmD from Streptomyces lavendulae NRRL 11002; SacD from Pseudomonas fluorescens A2–2; AZL_f00720 from Azospirillum sp. B510; KSE_08540 from Kitasatospora setae KM-6054.
FIGURE 7.
UV-Vis spectra (A) and MCD spectra (B) of SfmD wild type and mutants. SfmD WT (black solid), SfmD H191A (purple dot), SfmD H274A (green dot), SfmD H313A (red dot), SfmD C317A (blue dash).
DISCUSSION
Tyr and its derivatives not only play important physiological roles in living cells but also are precursors to many secondary metabolites with bioactivities. For example, l-Dopa, the aromatic ring hydroxylation product of Tyr, is marketed as a psychoactive drug for use in the clinical treatment of Parkinson's disease and dopamine-responsive dystonia. In addition, it is also an important precursor to the catecholamine neurotransmitters exemplified by dopamine and adrenaline (10) as well as the bioactive secondary metabolites exemplified by lincomycin (33) and CC-1065 (34). Several types of enzymes have been reported to catalyze the hydroxylation of the aromatic ring of Tyr or its derivatives, such as pteridine-dependent Tyr hydroxylase, copper-dependent tyrosinase, FAD-dependent SgcC, and others. A common feature of these enzymes is that all of them are oxygen-dependent non-heme-containing monooxygenases.
Recent reports have found that the non-proteinogenic amino acid precursor 3-OH-5-Me-OMe-Tyr originates from Tyr in the biosynthesis of SFM-A (6, 8). SfmD, a hypothetical protein thought to be responsible for the generation of the 3-hydroxy group (8), shows no homology to the known Tyr (or its derivatives) hydroxylases. Based on the UV-Vis spectra and LC-MS analysis of purified SfmD expressed in the absence or presence of ALA, we found that SfmD could bind heme as a cofactor. Moreover, the results of the pyridine hemochrome assays indicated that SfmD binds heme as a 1:1 complex. These findings confirmed that SfmD is a heme-containing protein that is different from the known Tyr (or its derivatives) hydroxylase.
Heme-containing proteins generally fall into two superfamilies: heme-containing oxygenases, which usually use oxygen as the oxidant, and heme-containing peroxidases, which usually use peroxide as the oxidant. No hydroxylation activity of SfmD was detected using the conditions reported for oxygenases with the plausible substrates in the presence of oxygen (see “Experimental Procedures”). To our surprise, SfmD showed some catalase activity that could dismutate H2O2. Moreover, in the presence of H2O2, SfmD could catalyze the hydroxylation of 3-Me-Tyr to 3-OH-5-Me-Tyr. Further analysis using an H218O2 assay confirmed that the oxygen inserted into the product originated from H2O2. These data indicate that SfmD is a H2O2-dependent heme-containing protein. As reported in the literature, there are two types of heme-containing proteins that are H2O2-dependent: heme-containing peroxidases and H2O2-dependent P450s. To determine the type that SfmD belongs to, the classical carbon monoxide (CO) binding experiments (35) were carried out. As shown in supplemental Fig. S9 and Table S3, the Soret band of SfmD changed to 418 nm, not ∼450 nm, after treatment with Na2S2O4 and CO, which ruled out the H2O2-dependent P450 family of proteins. Moreover, SfmD showed properties similar to the heme-containing peroxidase in low temperature EPR studies. Therefore, we concluded that SfmD is a heme-containing peroxidase that catalyzes the hydroxylation of Tyr or its derivatives using H2O2 as the oxidant.
Heme-containing peroxidases are widely distributed among bacteria, archaea, and eukarya, and catalyze one- and two-electron oxidation reactions of a great diversity of inorganic and organic compounds with H2O2. All currently available gene sequences of these metalloenzymes (listed in the PeroxiBase website) can be phylogenetically divided into two superfamilies (peroxidase-cyclooxygenase and peroxidase-catalase superfamily) and three families (dyp-type peroxidases, heme-haloperoxidases and di-heme peroxidases) (36). Among these, heme-thiolate peroxidases, such as heme-thiolate haloperoxidases, and heme-thiolate aromatic peroxidases, were reported to possess the ability to transfer oxygen from peroxides to various organic substrates including aromatic compounds (37, 38). And also, horseradish peroxidase was reported to be effective in oxidizing Tyr to Dopa in the presence of dihydroxyfumaric acid and oxygen (39). But until now, none of the heme-containing peroxidases listed in PeroxiBase has been shown to catalyze the aromatic ring hydroxylation of Tyr (or its derivatives) in the presence of H2O2. SfmD shows no homology to these heme-containing peroxidases listed in the PeroxiBase, and is a novel heme-containing peroxidase reported to catalyze the aromatic ring hydroxylation of Tyr or its derivatives using H2O2 as the oxidant. Very recently, Orf13 involved in the biosynthesis of anthramycin was also reported to be a heme-containing peroxidase catalyzing the hydroxylation of Tyr to Dopa in the presence of H2O2 around the time we submitted this article (40). And Orf13 also had the ability to catalyze the hydroxylation of Tyr to Dopa by a molecular oxygen-dependent pathway in the presence of dihydroxyfumaric acid or ascorbic acid (40). But SfmD showed no hydroxylation activity under the same conditions as reported for Orf13 using oxygen as the oxidant (data not shown). And meanwhile, SfmD could dismutate H2O2, while Orf13 would be inactivated when only reacted with H2O2. In addition, these two proteins show no homology to each other. All these indicate that distinct from Orf13 SfmD belongs to another type of heme-containing peroxidase.
Among the plausible substrates, both Tyr and 3-Me-Tyr can be hydroxylated by SfmD. Kinetic study results indicated that the preferred substrate of SfmD is 3-Me-Tyr. In combination with the assay results from SfmM2, we ascertained the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr. As shown in Fig. 1C, Tyr is methylated to 3-Me-Tyr, followed by SfmD-mediated hydroxylation to 3-OH-5-Me-Tyr, and finally transformed to 3-OH-5-Me-OMe-Tyr. Moreover, three similar proteins, SacD/SacF/SacG, were also found to be involved in the biosynthesis of SAC-B. In this study, we also characterized SacD as a SfmD-like heme-containing peroxidase. Together with the assay results from C-methyltransferase SacF, where only Tyr can be methylated,4 the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr involved in the biosynthesis of SAC-B was found to be identical to the aforementioned pathway (Fig. 1C). In addition, it has been shown that naphthyridinomycin, which possesses the same core quinone structure as SFM-A, can incorporate Tyr, 3-Me-Tyr, and 3-OH-5-Me-Tyr, but not Dopa (5). These data are consistent with our results that were characterized in vitro. Together, these findings indicate that the SfmD-like heme-containing peroxidase may be commonly involved in the biosynthesis of tetrahydroisoquinoline family members that feature the same core quinone structure with SFM-A, and imply that the biosynthetic pathway of 3-OH-5-Me-OMe-Tyr might be similar in the biosynthesis of these members.
In addition to SacD, there are two other homologs of SfmD in the NCBI data base, AZL_f00720 and KSE_08540, both of which are hypothetic proteins from genome sequences. Both are all located in putative secondary metabolite gene clusters, although the functions are yet to be established. A sequence alignment of SfmD with its analogs identified three conserved motifs within the C-terminal domain. Mutation studies confirmed that the conserved motif HXXXC was crucial for heme binding. This motif is different from the known heme binding motifs reported in other heme-containing proteins, such as FXXGXXXCXG, CXXCH, HPGG, CP, and GXXDG, which suggests that SfmD and its analogs may belong to a novel type of heme-containing protein family. In addition, these SfmD mutants lost the ability to hydroxylate 3-Me-Tyr, suggesting that the other two conserved motifs also play important roles in the catalytic cycle. However, the actual functions of these motifs in the catalytic cycle are still unknown and will require additional structural investigations.
In conclusion, we found that SfmD, along with its analog SacD, are clearly heme-containing peroxidases that catalyze the regioselective hydroxylation of 3-Me-Tyr to 3-OH-5-Me-Tyr using H2O2 as the oxidant. These findings not only have significance for the biosynthesis of other tetrahydroisoquinoline family members that possess the same core quinone structure with SFM-A, but also set the stage for elucidation of the mechanism for these newfound heme-containing peroxidases.
Acknowledgments
We thank Prof. Ben Shen, Departments of Chemistry and Molecular Therapeutics and Natural Products Library Initiative at The Scripps Research Institute, and Dr. Xu-Dong Qu of Shanghai Institute of Organic Chemistry for valuable advice and suggestions. We also thank Prof. Jian-Hua Ju and Dr. Bo Wang of South China Sea Institute of Oceanology, Chinese Academy of Sciences to help us order H218O2; Prof. Zi-Xin Deng' Laboratory of Shanghai JiaoTong University for support in obtaining MS data of proteins; Prof. Yu-Heng Zhang and Wei Tong of High Magnetic Field Laboratory, Chinese Academy of Science for assistance with EPR analysis.
This work was supported in part by grants from the National Basic Research Program of China (973 Program) (2010CB833200 and 2012CB721100), the National Natural Science Foundation of China (20832009 and 20921091), and the Chinese Academy of Science (KJCX2-YW-H08).

This article contains supplemental Figs. S1–S9 and Tables S1–S3.
G. Tang, unpublished data.
- SFM-A
- Saframycin A
- ALA
- 5-aminolevulinic acid
- Dopa
- 3,4-dihydroxyphenylalanine
- EPR
- electron paramagnetic resonance
- ESI-MS
- electrospray ionization-mass spectrometry
- ET-743
- Ecteinascidin 743
- HPLC
- high performance liquid chromatography
- IDO
- indoleamine 2,3-dioxygenase
- IPTG
- isopropyl β-D-thiogalactopyranoside
- MCD
- magnetic circular dichroism
- 3-Me-Tyr
- 3-methyltyrosine
- 3-Me-OMe-Tyr
- 3-methyl-O-methyltyrosine
- 3-OH-5-Me-Tyr
- 3-hydroxy-5-methyltyrosine
- 3-OH-5-Me-OMe-Tyr
- 3-hydroxy-5-methyl-O-methyltyrosine
- OMe-Tyr
- O-methyltyrosine
- SAC-B
- Safracin B
- SacD
- a putative hydroxylase involved in the biosynthesis of safracin B
- SAM
- S-adenosylmethionine
- SfmM2
- a putative C-methyltransferase involved in the biosynthesis of saframycin A
- TCA
- trichloroacetic acid
- TDO
- tryptophan 2,3-dioxygenase.
REFERENCES
- 1. Scott J. D., Williams R. M. (2002) Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem. Rev. 102, 1669–1730 [DOI] [PubMed] [Google Scholar]
- 2. Arai T., Takahashi K., Kubo A. (1977) New antibiotics saframycins A, B, C, D, and E. J. Antibiot. 30, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 3. Cuevas C., Francesch A. (2009) Development of Yondelis (trabectedin, ET-743). A semisynthetic process solves the supply problem. Nat. Prod. Rep. 26, 322–337 [DOI] [PubMed] [Google Scholar]
- 4. Mikami Y., Takahashi K., Yazawa K., Arai T., Namikoshi M., Iwasaki S., Okuda S. (1985) Biosynthetic studies on saframycin A, a quinone antitumor antibiotic produced by Streptomyces lavendulae. J. Biol. Chem. 260, 344–348 [PubMed] [Google Scholar]
- 5. Palaniswamy V. A., Gould S. J. (1986) The incorporation of 3′-methyltyrosine and 5′-methyl DOPA into naphthyridinomycin. J. Am. Chem. Soc. 108, 5651–5652 [Google Scholar]
- 6. Li L., Deng W., Song J., Ding W., Zhao Q.-F., Peng C., Song W.-W., Tang G.-L., Liu W. (2008) Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J. Bacteriol. 190, 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Velasco A., Acebo P., Gomez A., Schleissner C., Rodríguez P., Aparicio T., Conde S., Muñoz R., de la Calle F., Garcia J. L., Sánchez-Puelles J. M. (2005) Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2–2: designing new cytotoxic compounds. Mol. Microbiol. 56, 144–154 [DOI] [PubMed] [Google Scholar]
- 8. Fu C. Y., Tang M. C., Peng C., Li L., He Y. L., Liu W., Tang G. L. (2009) Biosynthesis of 3-hydroxy-5-methyl-o-methyltyrosine in the saframycin/ safracin biosynthetic pathway. J. Microbiol. Biotechnol. 19, 439–446 [DOI] [PubMed] [Google Scholar]
- 9. Koketsu K., Watanabe K., Suda H., Oguri H., Oikawa H. (2010) Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat. Chem. Biol. 6, 408–410 [DOI] [PubMed] [Google Scholar]
- 10. Fitzpatrick P. F. (1999) Tetrahydropterin-dependent amino acid hydroxylases. Ann. Rev. Biochem. 68, 355–381 [DOI] [PubMed] [Google Scholar]
- 11. Sánchez-Ferrer A., Rodríguez-López J. N., Garcia-Cánovas F., García-Carmona F. (1995) Tyrosinase: a comprehensive review of its mechanism. Biochim. Biophys. Acta 1247, 1–11 [DOI] [PubMed] [Google Scholar]
- 12. Lin S., Van Lanen S. G., Shen B. (2008) Characterization of the two-component, FAD-dependent monooxygenase SgcC that requires carrier protein-tethered substrates for the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Am. Chem. Soc. 130, 6616–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd ed., Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 14. Trost B. M., Rudd M. T. (2003) Chemoselectivity of the ruthenium-catalyzed hydrative diyne cyclization: total synthesis of (+)-cylindricine C, D, and E. Org. Lett. 5, 4599–4602 [DOI] [PubMed] [Google Scholar]
- 15. Deboves H. J., Montalbetti C. A., Jackson R. F. (2001) Direct synthesis of Fmoc-protected amino acids using organozinc chemistry: application to polymethoxylated phenylalanines and 4-oxoamino acids. J. Chem. Soc. Perkin Trans. 1, 1876–1884 [Google Scholar]
- 16. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 17. Berry E. A., Trumpower B. L. (1987) Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 [DOI] [PubMed] [Google Scholar]
- 18. Chowdhury G., Murayama N., Okada Y., Uno Y., Shimizu M., Shibata N., Guengerich F. P., Yamazaki H. (2010) Human liver microsomal cytochrome P450 3A enzymes involved in thalidomide 5-hydroxylation and formation of a glutathione conjugate. Chem. Res. Toxicol. 23, 1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alberta J. A., Dawson J. H. (1987) Purification to homogeneity and initial physical characterization of secondary amine monooxygenase. J. Biol. Chem. 262, 11857–11863 [PubMed] [Google Scholar]
- 20. Tenhunen R., Marver H. S., Schmid R. (1968) The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc. Natl. Acad. Sci. U.S.A. 61, 748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith W. L., Marnett L. J. (1991) Prostaglandin endoperoxide synthase: structure and catalysis. Biochim. Biophys. Acta 1083, 1–17 [DOI] [PubMed] [Google Scholar]
- 22. Sheoran A., King A., Velasco A., Pero J. M., Garneau-Tsodikova S. (2008) Characterization of TioF, a tryptophan 2,3-dioxygenase involved in 3-hydroxyquinaldic acid formation during thiocoraline biosynthesis. Mol. Biosyst. 4, 622–628 [DOI] [PubMed] [Google Scholar]
- 23. Huff A. M., Chang C. K., Cooper D. K., Smith K. M., Dawson J. H. (1993) Imidazole- and alkylamine-ligated iron (II, III) chlorin complexes as models for histidine and lysine coordination to iron in dihydroporphyrin-containing proteins: characterization with magnetic circular dichroism spectroscopy. Inorg. Chem. 32, 1460–1466 [Google Scholar]
- 24. Sugano Y. (2009) DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life Sci. 66, 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sono M., Roach M. P., Coulter E. D., Dawson J. (1996) Heme-containing Oxygenases. Chem. Rev. 96, 2841–2888 [DOI] [PubMed] [Google Scholar]
- 26. Poulos T. L. (2010) Thirty years of heme peroxidase structural biology. Arch. Biochem. Biophys. 500, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palmer G. (1983) in Iron Porphyrins: Part II (Lever A. B. P., Gray H. B., eds) pp. 43–88, Addison-Wesley, Reading, MA [Google Scholar]
- 28. Khindaria A., Aust S. D. (1996) EPR detection and characterization of lignin peroxidase porphyrin pi-cation radical. Biochemistry 35, 13107–13111 [DOI] [PubMed] [Google Scholar]
- 29. Khatri Y., Hannemann F., Ewen K. M., Pistorius D., Perlova O., Kagawa N., Brachmann A. O., Müller R., Bernhardt R. (2010) The CYPome of Sorangium cellulosum So ce56 and identification of CYP109D1 as a new fatty acid hydroxylase. Chem. Biol. 17, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 30. Allen J. W., Leach N., Ferguson S. J. (2005) The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem. J. 389, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guillou H., D'Andrea S., Rioux V., Barnouin R., Dalaine S., Pedrono F., Jan S., Legrand P. (2004) Distinct roles of endoplasmic reticulum cytochrome b5 and fused cytochrome b5-like domain for rat δ6-desaturase activity. J. Lipid Res. 45, 32–40 [DOI] [PubMed] [Google Scholar]
- 32. Mense S. M., Zhang L. (2006) Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 16, 681–692 [DOI] [PubMed] [Google Scholar]
- 33. Neusser D., Schmidt H., Spizèk J., Novotnà J., Peschke U., Kaschabeck S., Tichy P., Piepersberg W. (1998) The genes lmbB1 and lmbB2 of Streptomyces lincolnensis encode enzymes involved in the conversion of l-tyrosine to propylproline during the biosynthesis of the antibiotic lincomycin A. Arch. Microbiol. 169, 322–332 [DOI] [PubMed] [Google Scholar]
- 34. Hurley L. H., Rokem J. S. (1983) Biosynthesis of the antitumor antibiotic CC-1065 by Streptomyces zelensis. J. Antibiot. 36, 383–390 [DOI] [PubMed] [Google Scholar]
- 35. Hannemann F., Bichet A., Ewen K. M., Bernhardt R. (2007) Cytochrome P450 systems-biological variations of electron transport chains. Biochim. Biophys. Acta 1770, 330–344 [DOI] [PubMed] [Google Scholar]
- 36. Zamocky M., Obinger C. (2010) in Biocatalysis Based on Heme Peroxidases, (Torres E., Ayala M., eds) Springer-Verlag, Berlin, Heidelberg [Google Scholar]
- 37. Hofrichter M., Ullrich R. (2006) Heme-thiolate haloperoxidases: versatile biocatalysts with biotechnological and environmental significance. Appl. Microbiol. Biotechnol. 71, 276–288 [DOI] [PubMed] [Google Scholar]
- 38. Hofrichter M., Ullrich R., Pecyna M. J., Liers C., Lundell T. (2010) New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87, 871–897 [DOI] [PubMed] [Google Scholar]
- 39. Smith P. I., Swan G. A. (1976) A study of the supposed hydroxylation of tyrosine catalysed by peroxidase. Biochem. J. 153, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Connor K. L., Colabroy K. L., Gerratana B. (2011) A heme peroxidase with a functional role as an l-tyrosine hydroxylase in the biosynthesis of anthramycin. Biochemistry. 50, 8926–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]