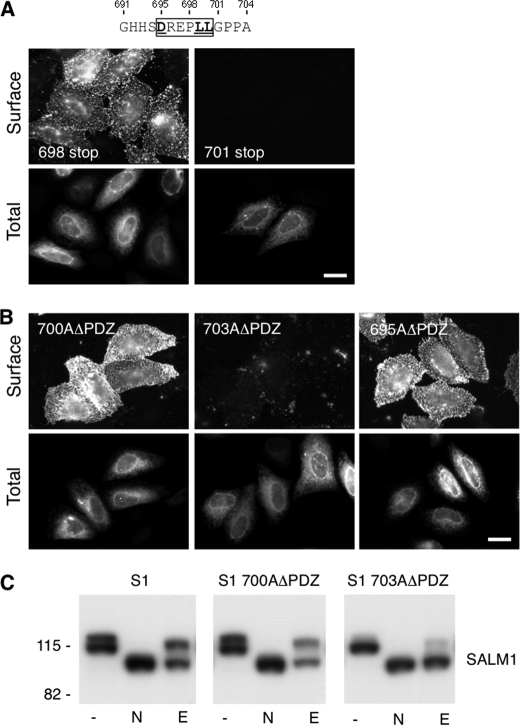
FIGURE 4.
Identification of a SALM1 dileucine ER retention motif. A, two additional Myc-SALM1 deletion constructions (698 stop and 701 stop) were made to further isolate the retention motif. Myc-SALM1 truncations (698 stop or 701 stop) were transfected into HeLa cells, and surface staining was detected by anti-S1NT (Surface). Cells were fixed and permeabilized to detect intracellular SALM1 with anti-Myc antibodies (Total). B, mutagenesis was used to mutate Leu-700 and Pro-703 to alanine in Myc-SALM1ΔPDZ (L700AΔPDZ and P703AΔPDZ, respectively), and both surface and total staining of transfected cells was performed. Scale bar, 20 μm. C, lysate from HeLa cells transfected with Myc-SALM1, Myc-SALM1 L700AΔPDZ, or Myc-SALM1 P703AΔPDZ was left untreated (−) or digested with N-glycosidase (N) or Endo H (E). All digests were followed by SDS-PAGE, and SALM1 was detected by immunoblotting with a polyclonal anti-S1NT antibody. Both mutant and WT proteins show more rapid migration following glycosidase cleavage of N-linked oligosaccharides, whereas only SALM1 P703AΔPDZ is fully digested by the high mannose-directed glycosidase Endo H. Molecular mass markers are shown in kDa.