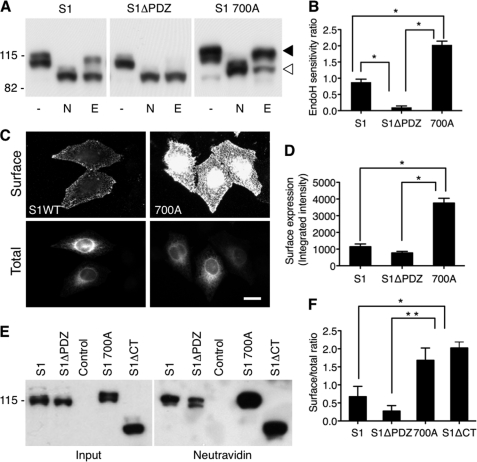
FIGURE 5.
Regulation of SALM1 ER retention and surface expression. A, lysate from HeLa cells transfected with Myc-SALM1, Myc-SALM1ΔPDZ, or Myc-SALM1 L700A was left untreated (−) or digested with N-glycosidase (N) or Endo H (E). Expression of SALM1 constructs was detected by immunoblotting with a polyclonal anti-S1NT antibody. SALM1 L700A is primarily Endo H-insensitive compared with SALM1 (top band, filled arrowhead), whereas most of the SALM1ΔPDZ expressed is Endo H-sensitive (bottom band, open arrowhead), suggesting that it is ER-retained. B, quantification of the ratio of Endo H-insensitive to Endo H-sensitive fraction using a densitometer indicates that SALM1 L700A is significantly less retained in the ER. Histograms show mean ± S.E. (error bars) (S1, 0.88 ± 0.09, n = 6; S1ΔPDZ, 0.10 ± 0.04, n = 6; 700A, 2.03 ± 0.08, n = 5; *, p < 0.05, one-way ANOVA). C, comparison of wild type Myc-SALM1 with Myc-SALM1 L700A indicates that mutation of residue Leu-700 enhances surface expression of SALM1. Surface staining was detected by anti-S1NT (Surface), and total SALM1 was detected using anti-Myc antibody (Total). D, quantification of surface expression of Myc-SALM1, Myc-SALM1ΔPDZ, and Myc-SALM1 L700A using laser scanning cytometry shows a significant increase in surface expression of SALM1 L700A. Histograms show mean ± S.E. (S1, 1145 ± 161, n = 4; S1ΔPDZ, 771 ± 92, n = 4; 700A, 3754 ± 289, n = 4; one-way ANOVA, p < 0.001). E, HeLa cells transfected with Myc-SALM1, Myc-SALM1ΔPDZ, control pCDNA3.1 vector, Myc-SALM1 L700A, or Myc-SALM1ΔCT were labeled with biotin, washed with glycine, and then lysed. Surface-labeled SALM was precipitated with NeutrAvidin beads. SALM1 constructs were detected by immunoblotting with a polyclonal anti-S1NT antibody. F, quantification of the surface (NeutrAvidin pull-down)/total (input) ratio using a densitometer. Histograms show mean ± S.E. (S1, 0.53 ± 0.33, n = 3; S1ΔPDZ, 0.24 ± 0.17, n = 3; 700A, 1.68 ± 0.34, n = 3; S1ΔCT, 2.02 ± 0.17, n = 3; *, p < 0.05; **, p < 0.001, one-way ANOVA).