Background: Recent proteomic studies suggest that lysine acetylation might regulate many metabolic pathways.
Results: NAD+-dependent deacetylase Sirt1 deacetylates phosphoglycerate mutase-1 (PGAM1) and attenuates catalytic activity.
Conclusion: Sirt1 directly modulates glycolysis.
Significance: These results advance our understanding of the regulatory impact of reversible acetylation on major metabolic pathways.
Keywords: Glycolysis, Histone Deacetylase, Metabolic Regulation, Post-translational Modification, Sirtuins, NAD+, Acetylation, Phosphoglycerate Mutase
Abstract
Emerging proteomic evidence suggests that acetylation of metabolic enzymes is a prevalent post-translational modification. In a few recent reports, acetylation down-regulated activity of specific enzymes in fatty acid oxidation, urea cycle, electron transport, and anti-oxidant pathways. Here, we reveal that the glycolytic enzyme phosphoglycerate mutase-1 (PGAM1) is negatively regulated by Sirt1, a member of the NAD+-dependent protein deacetylases. Acetylated PGAM1 displays enhanced activity, although Sirt1-mediated deacetylation reduces activity. Acetylation sites mapped to the C-terminal “cap,” a region previously known to affect catalytic efficiency. Overexpression of a constitutively active variant (acetylated mimic) of PGAM1 stimulated flux through glycolysis. Under glucose restriction, Sirt1 levels dramatically increased, leading to PGAM1 deacetylation and attenuated activity. Previously, Sirt1 has been implicated in the adaptation from glucose to fat burning. This study (i) demonstrates that protein acetylation can stimulate metabolic enzymes, (ii) provides biochemical evidence that glycolysis is modulated by reversible acetylation, and (iii) demonstrates that PGAM1 deacetylation and activity are directly controlled by Sirt1.
Introduction
Metabolic processes are regulated by a number of different mechanisms, including allosteric regulation, enzyme degradation, and reversible phosphorylation. Recently, reversible acetylation of protein lysine residues has been shown to directly regulate enzymatic activity, including acetyl-CoA synthetase 1 and 2, oxidative phosphorylation complex I, glutamate dehydrogenase, carbamoyl-phosphate synthase 1 (CPS1), long chain acyl-CoA dehydrogenase, HMG-CoA synthase 2, isocitrate dehydrogenase 2 (IDH2), superoxide dismutase 2 (SOD2), and phosphoenolpyruvate carboxykinase (1–12). Except for yeast phosphoenolpyruvate carboxykinase (4), acetylation decreased enzymatic activity, although the molecular mechanism has not been resolved in the case of phosphoenolpyruvate carboxykinase. Supporting the idea that acetylation is a widespread post-translational modification, a recent study revealed that most of the enzymes in glycolysis and gluconeogenesis display acetylated lysine residues (13). Although recent proteomic studies have identified hundreds of acetylated proteins, the cellular and molecular functions of these acetylations are >99% unknown (13–15). Understanding the molecular details of acetylation is essential to uncover the functional importance of these post-translational modifications and their implication for modulating a myriad of cellular processes.
One enzyme of particular interest, phosphoglycerate mutase-1 (PGAM1, EC 5.4.2.1), was identified in three separate acetyl proteomic screens (13–15). PGAM1 catalyzes the interconversion of 3-phosphoglycerate (3-PGA)2 to 2-phosphoglycerate (2-PGA), which includes the eighth step in glycolysis (16). PGAM1 becomes activated or “primed” by an intrinsic phosphatase activity that converts 2,3-bisphosphoglycerate to 2-PGA, phosphorylating the active site histidine in the process (16). The phosphohistidine is essential to carryout the mutase reaction, converting 3-PGA to 2-PGA in glycolysis. Historically, PGAM1 was not thought to catalyze a rate-determining step in glycolysis, but interestingly, PGAM1 is the rate-limiting enzyme of glycolysis in tumor cells, heart tissue, and leukocytes (17, 18). Inhibition of PGAM1 by novel epoxide inhibitors is lethal to cancer cells (19, 20). Taken together, these observations place PGAM at a critical regulatory step in glycolysis and provide insight into the Warburg effect in cancer cells. The possibility that PGAM1 and glycolysis are regulated by reversible acetylation has widespread implications in energy metabolism, metabolic syndromes, and cancer.
The sirtuins (NAD+-dependent deacetylases) have emerged as an important class of enzymes that deacetylates a growing list of non-histone proteins (1). Sirtuins catalyze the NAD+-dependent deacetylation of protein-acetylated lysines, catabolizing NAD+ in the process. Sirtuins have been implicated as central regulators in delaying metabolic diseases and the aging process (21, 22). The seven mammalian sirtuins (Sirt1–7) display distinct subcellular localization patterns (23) and regulate metabolism and other cellular processes, including genome maintenance and apoptosis (1, 24). Notably, Sirt1 is linked to lipid metabolism, fat storage, and fatty acid oxidation by regulation of PPARγ, to mitochondrial biogenesis by PGC-1α, and to insulin secretion by mediating UCP2 and PTP1B expression (25–28).
In this study, we provide detailed cellular and in vitro evidence for the direct regulation of glycolysis by sirtuin-mediated deacetylation of PGAM1. Prior to this study, detailed investigations of several metabolic enzymes indicated that acetylation inhibits activity. Here, we show that Sirt1-dependent deacetylation of PGAM1 reduces the rate of catalysis. The critical acetylation sites were mapped to the C-terminal regulatory cap of PGAM. Importantly, these lysines are novel acetylation sites that were not observed in prior MS studies, likely as a result of technical limitations detecting very short peptides. Glucose concentrations were sufficient to regulate Sirt1 protein levels. Under glucose restriction, Sirt1 proteins levels increase dramatically (11 fold), enabling Sirt1-mediated deacetylation of PGAM1 and restricted flux through glycolysis.
EXPERIMENTAL PROCEDURES
Materials
The plasmid pcDNA3.1 encoding Sirt1–7 with a FLAG tag was obtained from Dr. E. Verdin at the University of California (San Francisco). The plasmid pCVM-1 encoding PGAM1 was purchased from Origene. Antibodies used in the study were anti-acetyl-lysine polyclonal (Cell Signaling), anti-PGAM1 (Abcam), anti-β-actin (clone AC-15, Sigma), and anti-FLAG (M2 Sigma).
Cell Culture and Transfection
HEK293 cells were cultured as a monolayer in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum. Co-transfection of HEK293 cells was performed using 4 mg of total DNA or 50 pmol of siRNA from Dharmacon with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol and as described in the figure legends. Cell treatments were carried out as described in the figure legends. Soluble cell extracts were made in RIPA buffer (Sigma) with 5 mm nicotinamide, 100 μm TSA, and HALT protease inhibitor (Pierce). Protein concentration was quantified by Bradford assay (Bio-Rad).
Immunoprecipitation and Western Blotting of PGAM1
Immunoprecipitations were performed with anti-MYC antibody-conjugated agarose resin and anti-PGAM1 antibody with protein G conjugates as described by the manufacturer. Western blotting and SDS-PAGE were carried out under standard practice with PVDF membranes (29).
Expression and Purification of Recombinant Sirt1
Sirt1 from mammalian expression vectors was cloned into the bacterial expression vector pQE-80 (Qiagen). The plasmid was transformed into Escherichia coli BL21DE3. The transformed bacteria were induced with isopropyl d-thiogalactopyranoside, lysed, and purified by nickel affinity chromatography.
PGAM1 Activity Assay
The forward PGAM1 activity was determined by coupling the formation of 2-phosphoglycerate from 3-phosphoglycerate with the enolase-, pyruvate kinase-, and lactate dehydrogenase-catalyzed reactions. Assays were performed at 37 °C and measured at 340 nm according to Winn et al. (28). Cell lysates for enzymatic activity were prepared with lysis buffer. The reaction mixture contained 100 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 2 mm MgCl2, 100 mm KCl, 0.2 mm NADH, 1.5 mm ADP, 10 μm 2,3-bisphosphoglycerate, lactate dehydrogenase (0.6 unit/ml), pyruvate kinase (0.5 unit/ml), enolase (0.3 unit/ml), and 1 mm 3-phosphoglycerate. PGAM1 reverse activity was determined by coupling the formation of 3-phosphoglycerate from 2-phosphoglycerate with the phosphoglycerate kinase and glyceraldehyde phosphate dehydrogenase-catalyzed reactions. The assay mixture contained 100 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 2 mm MgCl2, 100 mm KCl, 0.2 mm NADH, 1.5 mm ADP, 10 μm 2,3-bisphosphoglycerate, 1 mm 2-phosphoglycerate, 3.3 units of glyceraldehyde phosphate dehydrogenase, and 2 units of phosphoglycerate kinase. Assays were performed at 25 °C and measured at 340 nm. PGAM activity was measured in the linear range of initial velocity, and no more than 10% product was formed. Use of catalytic amounts of PGAM was established by ensuring that the observed rate was linear with respect to PGAM levels (in purified form or from cell extracts). Experiments were performed at least three times in triplicate.
Sirt1 Deacetylation Assay
Deacetylation of acetylated PGAM1 by Sirt1 was performed in 100 ml of Tris, pH 7.5, 1 mm NAD+, and 1 mm DTT at 37 °C (30). Specific assay conditions are indicated in the figure legends.
Metabolite Analysis
Transfected cells were cultured in regular DMEM with 10% (v/v) FBS lacking pyruvate by standard methods. Cell culture supernatants were collected and immediately frozen in liquid nitrogen. Glucose, lactate, and alanine were measured in the thawed supernatants as described previously (31).
RESULTS
Sirtuin Inhibitor Nicotinamide Promotes PGAM1 Acetylation
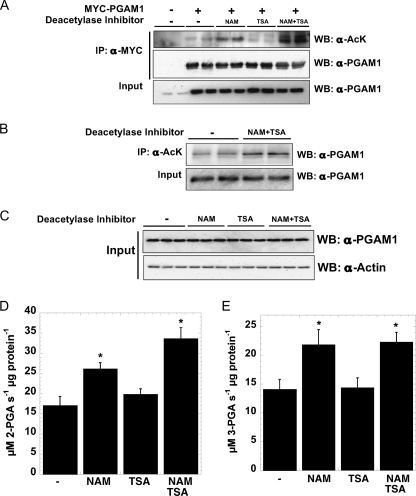
To assess the acetylation dynamics of PGAM1, HEK293 cells transiently expressing PGAM1 were treated with protein deacetylase inhibitors, 10 mm nicotinamide (NAM) or 5 μm TSA, or both. Following treatment, PGAM1 was immunoprecipitated, and the acetylation state was probed using Western blot analysis with an anti-acetyl antibody (Fig. 1A). Cells treated with NAM, an inhibitor of sirtuin-dependent deacetylation, revealed increased acetylation of PGAM1, whereas treatment with TSA alone led to insignificant changes in acetylation compared with mock treatment. Higher levels of acetylation were detected when cells were treated with both NAM and TSA. Examination of endogenous PGAM1 from HE329K cells revealed that acetylation was increased when cells were treated with NAM or the combination of NAM and TSA, but not TSA alone (supplemental Fig. 1). In the case of endogenous PGAM, NAM and TSA treatment did not lead to more acetylation than was detected by NAM alone. In a reciprocal experiment, HEK293 lysates were immunoprecipitated with an anti-acetyl-lysine antibody and then probed for levels of PGAM1. Higher levels (3 fold) of acetylated PGAM1 were present in cells treated with NAM and TSA, as compared with mock treatment (Fig. 1B). The ability of NAM to promote increased acetylation of PGAM1 suggested that sirtuin deacetylases regulate the acetylation status of PGAM1.
FIGURE 1.
PGAM1 is acetylated in vivo, and acetylation increases PGAM1 activity. A, transiently transfected PGAM1 is acetylated in vivo, when treated with sirtuin-specific inhibitors. MYC-PGAM1 was overexpressed in HEK293 cells and treated overnight with 5 mm NAM, 5 μm TSA, and 5 mm NAM, 5 μm TSA. MYC-PGAM1 was immunoprecipitated (IP) by anti-MYC conjugated to agarose at 4 °C for 4 h. These extracts were resolved by SDS-PAGE and detected by Western blotting (WB) with anti-acetyl-lysine (AcK) and anti-PGAM1 antibodies, respectively. Input refers to 1% of total lysate and was detected with anti-PGAM1 antibody. B, endogenous PGAM1 is acetylated in vivo. Endogenous PGAM1 is immunoprecipitated by anti-acetyl-lysine and protein G from HEK293 cells incubated overnight with 5 mm NAM, 5 μm TSA. These extracts (Input = 1% total cell lysate) were resolved by SDS-PAGE and detected by Western blotting with anti-PGAM1 antibody. C–E, soluble extracts of HEK293 cells incubated overnight with 5 mm NAM, 5 μm TSA, and 5 mm NAM, 5 μm TSA were resolved by SDS-PAGE and detected by Western blotting with anti-PGAM1 and anti-actin antibodies (C), respectively; activity assays measure PGAM1 activity. Input refers to 1% total cellular lysate. Forward PGAM reaction (D), coupled to Enolase, pyruvate kinase, and lactate dehydrogenase, utilized saturating 3-phosphoglycerate measuring absorbance at 340 nm. Reverse PGAM reaction (E), coupled to phosphoglycerate kinase and GAPDH, utilized saturating 3-phosphoglycerate measuring absorbance at 340 nm. Error bars represent S.E. (n = 3); *, p < 0.05 compared with untreated cells.
PGAM1 Acetylation Stimulates Enzymatic Activity
To assess the functional consequences of PGAM1 acetylation, HEK293 cells were treated with histone deacetylase inhibitors NAM, TSA, or NAM and TSA, and protein levels and activity of PGAM1 were determined (Fig. 1C). After 16 h of histone deacetylase inhibitor treatment, PGAM1 protein levels were unaffected (Fig. 1C). However, PGAM1 activity measurements revealed that NAM alone or combined NAM and TSA treatment led to a significant 30–40% increase (Fig. 1, D and E). With TSA treatment alone, PGAM1 activity was indistinguishable from that of the mock treatment (Fig. 1, D and E). The observation that both directions (3-PGA versus 2-PGA) of the PGAM1 reaction were similarly affected under saturating substrate concentrations suggests that catalysis (kcat) is the stimulated step. These results suggest that acetylation of PGAM1 stimulates activity and that deacetylation is controlled by a sirtuin deacetylase.
Sirt1-specific Deacetylation of PGAM1
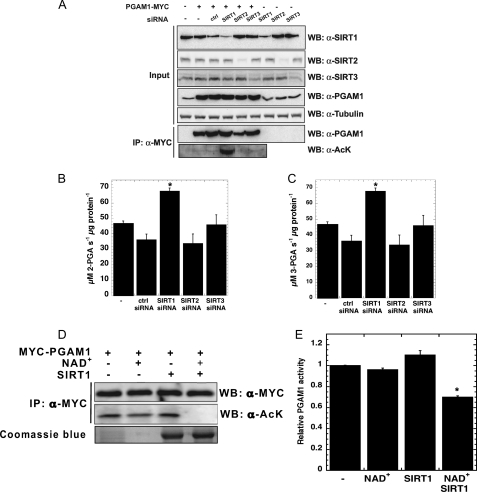
To identify the sirtuin(s) responsible for deacetylating PGAM1, we used siRNA knockdown directed at endogenous Sirt1, Sirt2, and Sirt3. Transfection of Sirt1, Sirt2, or Sirt3 siRNAs into HEK293 cells substantially reduced the endogenous protein levels of the individual sirtuins (Fig. 2A). When Sirt1 levels were lowered, there was a dramatic increase in PGAM1 acetylation, but no significant changes were observed in the control, Sirt2, or Sirt3 knockdown experiments. These findings indicate that among Sirt1, Sirt2, and Sirt3, only Sirt1 is capable of promoting deacetylation of PGAM1 in cultured cells.
FIGURE 2.
SIRT1 specifically deacetylates PGAM1. A, only SIRT1 knockdown significantly increased PGAM1-acetylated levels. HEK293 cells were transiently transfected with MYC-PGAM1 and siRNA to SIRT1–3. Soluble extracts were resolved by SDS-PAGE and detected through Western blot (WB), with anti-acetyl-lysine (AcK) and anti-PGAM1 antibodies. Input blots (1% total cellular extract) were detected with antibodies against PGAM1, SIRT1, SIRT2, SIRT3, and tubulin. B and C, activity was measured at 340 nm by PGAM1-coupled assay. D, acetylation of PGAM1 is decreased in vitro by SIRT1 deacetylation. Immunoprecipitants (IP) of MYC-PGAM1 from HEK293 cells transiently transfected with PGAM1 were incubated with the combination of NAD+ and SIRT1, and the reactions were resolved by SDS-PAGE and detected through Western blot, with anti-acetyl-lysine and anti-PGAM1 antibodies. E, activity was measured at 340 nm by PGAM1-coupled assay. Error bars represent S.E. (n = 3); *, p < 0.05 compared with control.
To assess the functional role of Sirt1-dependent deacetylation of PGAM1, the activity of PGAM was determined from cells treated with siRNAs of a mock control, Sirt1, Sirt2, and Sirt3 (Fig. 2, B and C). PGAM1 activity was 45% higher in the Sirt1 knockdown cells, compared with mock-treated, siRNA control, and Sirt2 and Sirt3 knockdown cells. To corroborate these findings, co-expressing studies were performed with all sirtuin proteins (including Sirt1–7). Specifically, HEK293 cells were transiently co-transfected with PGAM1 and the Sirt1–7 constructs. Cells were then lysed, and PGAM1 was immunoprecipitated overnight, and acetylation levels were probed using Western blot analysis. Among all seven sirtuins, cells expressing Sirt1 or Sirt2 showed a decrease in the acetylation of PGAM1 (supplemental Fig. 2A). We suspected that the PGAM1 deacetylation observed in Sirt2-overexpressing cells might be due to the high levels of Sirt2 overexpression and the long overnight incubation used for immunoprecipitation. To examine this, we measured PGAM1 activity immediately after cell lysis to minimize spurious deacetylation. In this experiment, expression of Sirt1, but not Sirt2 or Sirt3, decreased PGAM1 activity by 40–50% (supplemental Fig. 2, B and C). Together, these results suggest that Sirt1 modulates PGAM1 acetylation and activity.
Sirt1 Directly Deacetylates PGAM1 in Vitro
To decipher whether Sirt1 directly deacetylates PGAM1 or whether deacetylation of PGAM1 occurs by a secondary mechanism, acetylated PGAM1 was deacetylated in vitro. Specifically, hyperacetylation of PGAM1 was induced by treating HEK293 cells with NAM for 16 h. Acetylated PGAM1 was immunopurified and used as a substrate for Sirt1 (Fig. 2D). Addition of both NAD+ and purified recombinant Sirt1 was required for deacetylation of PGAM1, indicating that Sirt1 acts directly on PGAM1. Consistent with the cellular studies, deacetylation induced a decrease in catalytic efficiency of PGAM1 (Fig. 2E).
PGAM1 Is Acetylated at C-terminal Lys-251, Lys-253, and Lys-254
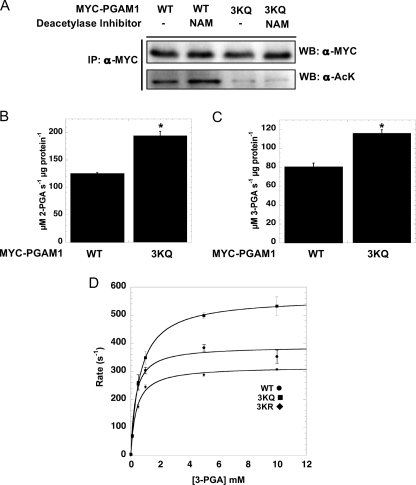
To determine the functionally important sites of Sirt1-dependent deacetylation on PGAM1, we evaluated a number of candidate lysine residues. From high throughput proteomic studies, the following lysines were reported to be acetylated: Lys-61, Lys-100, Lys-106, Lys-113, Lys-225, and Lys-228 (4, 14, 15). Each of these candidate sites was individually mutated to arginine (a deacetylated lysine mimic) or glutamine (an acetyl-lysine mimic), and the mutant proteins were expressed in cells, and PGAM1 acetylation levels were probed using Western blot analysis. Surprisingly, the site mutants displayed no significant change in the acetylation profile (data not shown). Based on these findings, it appeared that none of the residues previously identified by mass spectral proteomics comprise the regulatory site of PGAM1 acetylation suggested in this study. We next explored the possibility that the lysine regulatory site(s) might have been missed by proteomic analysis because the tryptic fragments might be too small for typically MS detection. The C terminus of PGAM1 harbors several adjacent lysine residues that include Lys-251, Lys-253, and Lys-254, and these would be invisible in the proteomic screens. This C-terminal region of PGAM has been hypothesized to affect enzyme efficiency by protecting the phosphohistidine intermediate within the activated form of the enzyme (32, 33). Mutation or deletion of this region led to decreased enzyme activity and increased hydrolysis of the phosphohistidine in the active site. To determine whether this region is responsible for the acetylation-dependent effect on activity, a triple mutant consisting of the lysine residues (Lys-251, Lys-253, and Lys-254) was constructed, and each residue was mutated to glutamine (referred to as 3KQ). Either wild type or the 3KQ mutant was expressed in HEK293 cells, and the acetylation status following NAM treatment was assessed by Western blot analysis (Fig. 3A). With the 3KQ mutant, there were negligible levels of basal acetylation, and NAM did not stimulate an increase compared with wild type PGAM1, which displayed both higher basal acetylation and a stimulated increase after NAM treatment (Fig. 3A). These results suggest that Lys-251, Lys-253, and Lys-254 are the primary acetylation sites, as they constitute >90% of the acetylation signal in Western analysis. PGAM1 activity assays were performed to determine whether the 3KQ mutant acted as partial surrogate for acetylated PGAM1. Indeed, comparison of catalytic activities indicated that the 3KQ mutant displayed an ∼40% higher rate compared with wild type PGAM1, suggesting that the 3KQ mutant, at least partially, mimics the effect of acetylated PGAM1 and that the C terminus of PGAM1 is the site of functionally relevant acetylation (Fig. 3, B and C). To corroborate these results, we constructed a 3KR mutant and compared the detailed steady-state kinetics between wild type, 3KQ, and 3KR mutants in vitro (Fig. 3D). The bacterially expressed and purified versions displayed similar Km values for 3-PGA (∼350 μm), but the 3KQ converted 3-PGA to 2-PGA with the highest kcat value of 563 (s−1), compared with 388 (s−1), for wild type and 318 (s−1) for 3KR mutant. These results are entirely consistent with the conclusion that acetylation stimulates PGAM activity through an increase in catalytic rate.
FIGURE 3.
C-terminal tail of PGAM1 is acetylated. A, MYC-PGAM1 and MYC-PGAM1 3KQ mutant (K251Q, K253Q, and K254Q) were transiently transfected into HEK293 cells following 5 mm NAM treated for 16 h. Cell lysates were immunoprecipitated (IP) with anti-MYC antibody, resolved by SDS-PAGE, and then detected with Western blot (WB) analysis with anti-acetyl-lysine (AcK) and anti-MYC antibodies. B and C, activity was measured at 340 nm by the PGAM1-coupled assay. D, PGAM1 3KQ shows high catalytic activity against 2-phosphoglycerate. Catalytic amounts of PGAM1 ●, PGAM1 3KQ ■, and PGAM1 3KR ⧫ were incubated with the indicated amounts of 3-PGA, and activity was measured at 340 nm by PGAM1-coupled assay. Data from the coupled assay were fitted to the Michaelis-Menten equation. Error bars represent S.E. (n = 3). *, p < 0.05.
Glucose Concentration Modulates Sirt1 Levels, PGAM1 Acetylation, and Glycolytic Flux
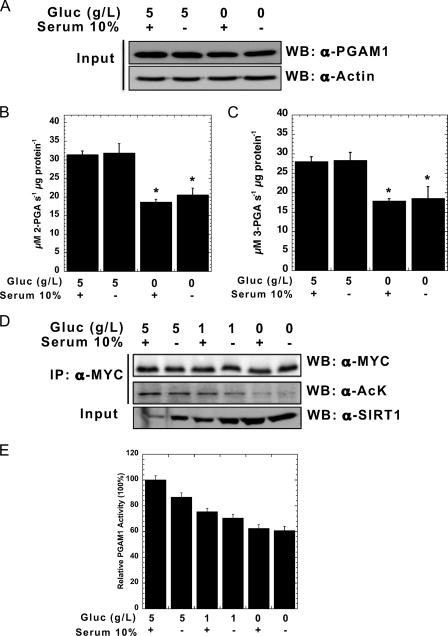
The presented data implicate Sirt1 as a negative regulator of PGAM1 activity, suggesting that Sirt1 might directly modulate glycolytic flux. To establish a physiological link between PGAM acetylation and glucose availability, we compared endogenous PGAM1 activities from HEK293 cells treated with either 0 or 5 g/liter glucose for 6 h. PGAM1 activity was nearly two fold higher in 5 g/liter glucose compared with 0 g/liter glucose (Fig. 4, A–C). To investigate further the link between glucose levels and PGAM acetylation, HEK293 cells overexpressing PGAM1 were grown in glucose-free DMEM supplemented with variable amounts of glucose and either in the presence or absence of serum (Fig. 4D). Western blot analysis was then used to probe the acetylation state of PGAM1, revealing that PGAM1 acetylation levels directly correlated with glucose concentration. Although the addition of serum had a slight enhancement of acetylation, serum was not essential for the general trend of increased acetylation as a function of glucose. To demonstrate that PGAM1 acetylation levels are qualitatively linked to activity, PGAM activities were measured under these six conditions (Fig. 4E). Consistent with our earlier findings that demonstrated acetylation promotes PGAM activity (Figs. 1–3), the extent of PGAM acetylation in response to different [glucose] was mirrored by an increase in PGAM activity (Fig. 4E). To provide a mechanism for the Sirt1-dependent regulation of PGAM, the levels of Sirt1 protein were determined under the six metabolic conditions (Fig. 4D). Dramatically, glucose restriction led to an 11-fold increase in Sirt1 protein levels (Fig. 4D), which was largely independent of serum treatment. The different levels of Sirt1 protein inversely correlated with PGAM acetylation. Collectively, these data suggest that glucose restriction increases Sirt1 protein levels, which leads to deacetylation and attenuation of PGAM activity.
FIGURE 4.
High glucose increases PGAM1 acetylation and activity in vivo. A, HEK293 cells were treated by 5 g/liter glucose (Gluc) ± FBS and 0 g/liter glucose ± FBS for 6 h. Endogenous PGAM1 activity assays were measured at 340 nm for both forward (B) and reverse reactions (C). Cell lysates were analyzed by SDS-PAGE and detected through Western blot (WB) with anti-PGAM1 and anti-actin antibodies. Input refers to 1% total cellular extract (A). D, MYC-PGAM1 was overexpressed in HEK293 cells following incubation for 6 h with 5 g/liter glucose ± FBS, 1 g/liter glucose ± FBS, and 0 g/liter glucose ± FBS. Cell lysates were resolved by SDS-PAGE, and Western blotting was performed with anti-SIRT1, anti-MYC, and anti-acetylated lysine antibodies. E, MYC-PGAM1 was immunoprecipitated (IP) with anti-MYC conjugated to agarose, and PGAM1 activity assay was measured. PGAM1 protein levels were normalized using quantification of the anti-MYC Western as in D. Input refers to 1% cellular extract. Error bars represent S.E. (n = 3); *, p < 0.05 compared with 5 g/liter glucose with serum.
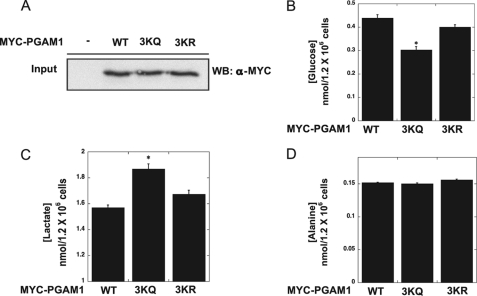
To provide evidence that the reversible acetylation of PGAM modulates glycolytic flux, we utilized the acetylation mimic 3KQ and the deacetylation mimic 3KR variants. We have demonstrated that the 3KQ mutant is more active than either wild type or the 3KR variant (Fig. 3), and thus it was hypothesized that overexpression of the 3KQ variant would increase glycolytic flux. Either 3KQ, 3KR, or wild type PGAM was overexpressed in HEK293 cells cultured in pyruvate-free media, and the concentrations of glucose, lactate, and alanine were measured. Growth in pyruvate-free media simplifies measurements of glycolytic flux (34). Compared with cells expressing PGAM and the 3KR variant, cells expressing 3KQ exhibited stimulated glucose consumption and a corresponding increase in lactate levels (Fig. 5, A–C). No significant changes in alanine levels were observed (Fig. 5D). These data demonstrate 3KQ can stimulate glycolysis, and together the results suggest that reversible acetylation of PGAM1 modulates activity and thus glycolytic flux.
FIGURE 5.
Expression of PGAM1 mutant 3KQ (acetylated mimic) stimulates glycolytic flux. WT MYC-PGAM1, 3KQ, and 3KR were overexpressed in HEK293 cells. A, input blots (1% total lysate) were detected with anti-MYC antibody. B, glucose levels in HEK293 cells expressing PGAM1, PGAM1 3KQ, and PGAM1 3KR. C, lactate levels in HEK293 cells expressing PGAM1, PGAM1 3KQ, and PGAM1 3KR. D, alanine levels in HEK293 cells expressing PGAM1, PGAM1 3KQ, and PGAM1 3KR. Error bars represent S.E. (n = 3); *, p < 0.05.
DISCUSSION
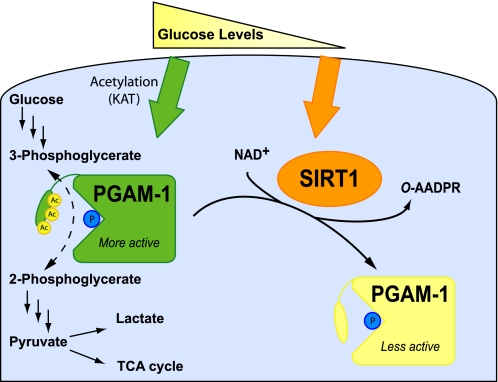
Emerging evidence indicates that reversible protein acetylation regulates multiple metabolic processes, including acetyl-CoA synthesis, urea cycle, REDOX status, and fatty acid oxidation (2–12). Here, we provide evidence for the direct regulation of glycolysis by sirtuin-mediated deacetylation of PGAM1. Importantly, we show that Sirt1-dependent deacetylation of PGAM1 reduces its catalytic activity. Prior to this study, detailed investigations of specific metabolic enzymes suggested that acetylation inhibits activity, and therefore deacetylation stimulates activity. Utilizing site-directed mutagenesis, and in vitro and cell-based analyses, Lys-251, Lys-253, and Lys-254 were identified as the regulatory acetylation sites. Furthermore, varied glucose concentrations were sufficient to regulate Sirt1 protein levels. Under glucose restriction, Sirt1 proteins levels increase dramatically, allowing Sirt1-mediated deacetylation of PGAM1 and restricted flux through glycolysis (Fig. 6).
FIGURE 6.
Model of PGAM1 regulation by glucose availability and SIRT1-dependent reversible deacetylation. Under high glucose, acetylation of the C-terminal cap of PGAM1 stimulates catalysis. Under glucose restriction, SIRT1 levels increase, leading to deacetylation of PGAM1 and decreased enzymatic activity.
The exact molecular details for enhanced catalysis by acetylated PGAM is unclear; however, the C-terminal region composed of Lys-251, Lys-253, and Lys-254 was previously proposed to act as a dynamic cap over the active site of PGAM1. Although PGAM structures are available, the C terminus is not visible, preventing direct structural insight. Biochemical data suggest that this cap protects the phosphohistidine intermediate and prevents hydrolysis, which would short circuit the catalytic cycle of the mutase reaction (32, 33). The C terminus of PGAM1 was hypothesized to interact with a phosphoglycerate substrate and position the substrate for catalysis (33). Supporting the important function of the cap, removing the C-terminal tail of PGAM1 in Saccharomyces cerevisiae decreased 10-fold the conversion of 3-PGA to 2-PGA (33), whereas the phosphatase activity (hydrolysis of the phosphohistidine intermediate) for this mutant was increased. Here, acetylated PGAM1 from cells displays enhanced catalysis, and in vitro the acetylation mimic 3KQ displays a similar increased rate of catalysis. Because the stoichiometry of acetylation on endogenous PGAM is unknown, and Lys-to-Gln substitutions cannot fully mimic an acetyl-lysine, the catalytic effect of complete acetylation might be more dramatic than is attainable under the conditions employed in our studies. Nevertheless, compared with wild type PGAM and 3KR, overexpression of the 3KQ was sufficient to increase flux through glycolysis. We hypothesize that acetylation of the C terminus optimizes the position of the cap to allow efficient phospho-transfer from the 3-position to the 2-position of glycerate, while protecting the enzyme intermediate from hydrolysis (Fig. 6).
Glucose restriction dramatically increases protein levels of Sirt1, which led to PGAM1 deacetylation and down-regulation of mutase activity. These findings are consistent with prior observations that showed Sirt1 protein is stabilized in cultured cells under nutrient deprivation (35). In addition to protein stabilization, increased NAD+ levels during low energy status are suggested to stimulate Sirt1 (21, 36). Two mechanisms have been proposed to explain increased NAD+ levels under conditions that promote Sirt1 activity. An increase in nicotinamide phosphoribosyltransferase expression stimulates synthesis of NAD+ and decreases the levels of nicotinamide, a potent sirtuin inhibitor (37, 38). When reducible substrates are scarce (e.g. low glucose), it has been proposed that nuclear-cytoplasmic NADH production is diminished, effectively increasing the pool of NAD+ (39). Although low energy input clearly induces a large increase in Sirt1 protein, the contributions from direct fluctuations in NAD+ and nicotinamide levels remain somewhat elusive.
This study provides the mechanistic evidence for the regulatory role of acetylation in glycolysis and the biochemical evidence for the direct involvement of NAD+-dependent protein deacetylases. Previously, Sirt1 was reported to affect metabolic pathways at the transcriptional level, promoting β-oxidation and inhibition of lipogenesis, as well as the modulation of gluconeogenesis (21). The function of Sirt1 in gluconeogenesis remains enigmatic, as there are reports suggesting either positive or negative regulatory control by Sirt1 (4, 40–43). Overall, these observations place Sirt1 at an important regulatory node of glucose homeostasis, and the apparent discrepancies might be explained through tissue-specific metabolism and differences in nutrient status of the cells. The emerging picture points to a modulatory role for Sirt1 during the transition from glucose utilization to fatty acid oxidation. The coordinate down-regulation of glycolysis would prevent the unnecessary depletion of glycolytic intermediates that are utilized in other biosynthetic pathways. Thus, PGAM1 deacetylation and down-regulation by Sirt1 would restrict the flow-through glycolysis and maintain an available 3-phosphoglycerate pool. 3-Phosphoglycerate is used to make serine, which serves as a precursor for glycine and cysteine, ultimately feeding into one-carbon metabolism that is essential for nucleic acid and phospholipid synthesis.
Examining the role of Sirt1-mediated regulation of PGAM1 in disease states, such as cancer metabolism and the impact on the Warburg effect, will likely provide novel insight into the contribution of dynamic acetylation (14). PGAM1 is considered an important target for developing cancer therapeutics, as PGAM1 inhibition is lethal to cancer cells in culture (20). Further work will be needed to explore whether Sirt1 dysfunction, which has been linked to many age-related afflictions, is propagated to dysregulation of glycolysis and a contributing factor to the Warburg effect (44).
Supplementary Material
Acknowledgment
We thank Jess Feldman for assistance during the preparation of this manuscript.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1 and 2.
- 3-PGA
- 3-phosphoglycerate
- PGAM
- phosphoglycerate mutase
- 2-PGA
- 2-phosphoglycerate
- TSA
- tricostatin A
- NAM
- nicotinamide.
REFERENCES
- 1. Smith B. C., Hallows W. C., Denu J. M. (2008) Mechanisms and molecular probes of sirtuins. Chem. Biol. 15, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hallows W. C., Lee S., Denu J. M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., Tao S. C., Qian J., Zhao Y., Boeke J. D., Berger S. L., Zhu H. (2009) Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qiu X., Brown K., Hirschey M. D., Verdin E., Chen D. (2010) Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12, 662–667 [DOI] [PubMed] [Google Scholar]
- 7. Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Someya S., Yu W., Hallows W. C., Xu J., Vann J. M., Leeuwenburgh C., Tanokura M., Denu J. M., Prolla T. A. (2010) Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009) SIRT5 deacetylates carbamoyl-phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 11. Shimazu T., Hirschey M. D., Hua L., Dittenhafer-Reed K. E., Schwer B., Lombard D. B., Li Y., Bunkenborg J., Alt F. W., Denu J. M., Jacobson M. P., Verdin E. (2010) SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 and regulates ketone body production. Cell Metab. 12, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hallows W. C., Yu W., Smith B. C., Devries M. K., Devires M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 15. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 16. Fothergill-Gilmore L. A., Watson H. C. (1989) The phosphoglycerate mutases. Adv. Enzymol. Relat. Areas Mol. Biol. 62, 227–313 [DOI] [PubMed] [Google Scholar]
- 17. Kashiwaya Y., Sato K., Tsuchiya N., Thomas S., Fell D. A., Veech R. L., Passonneau J. V. (1994) Control of glucose utilization in working perfused rat heart. J. Biol. Chem. 269, 25502–25514 [PubMed] [Google Scholar]
- 18. Shalom-Barak T., Knaus U. G. (2002) A p21-activated kinase-controlled metabolic switch up-regulates phagocyte NADPH oxidase. J. Biol. Chem. 277, 40659–40665 [DOI] [PubMed] [Google Scholar]
- 19. Evans M. J., Morris G. M., Wu J., Olson A. J., Sorensen E. J., Cravatt B. F. (2007) Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Mol. Biosyst. 3, 495–506 [DOI] [PubMed] [Google Scholar]
- 20. Evans M. J., Saghatelian A., Sorensen E. J., Cravatt B. F. (2005) Target discovery in small molecule cell-based screens by in situ proteome reactivity profiling. Nat. Biotechnol. 23, 1303–1307 [DOI] [PubMed] [Google Scholar]
- 21. Haigis M. C., Sinclair D. A. (2010) Mammalian sirtuins. Biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donmez G., Guarente L. (2010) Aging and disease. Connections to sirtuins. Aging Cell 9, 285–290 [DOI] [PubMed] [Google Scholar]
- 23. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu J., Auwerx J. (2009) The role of sirtuins in the control of metabolic homeostasis. Ann. N. Y. Acad. Sci. 1173, Suppl. 1, E10–E19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 27. Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E. J., Lin S. J., Guarente L. (2006) Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun C., Zhang F., Ge X., Yan T., Chen X., Shi X., Zhai Q. (2007) SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 6, 307–319 [DOI] [PubMed] [Google Scholar]
- 29. Harlow E., Lane D. (1988) Antibodies, A Laboratory Manual, pp. 471–510, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 31. Solanki K., Nyfeler F., Moser U. K., Walter P. (1980) Effect of glucose on carbohydrate synthesis from alanine or lactate in hepatocytes from starved rats. Biochem. J. 192, 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winn S. I., Watson H. C., Harkins R. N., Fothergill L. A. (1981) Structure and activity of phosphoglycerate mutase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 293, 121–130 [DOI] [PubMed] [Google Scholar]
- 33. Walter R. A., Nairn J., Duncan D., Price N. C., Kelly S. M., Rigden D. J., Fothergill-Gilmore L. A. (1999) The role of the C-terminal region in phosphoglycerate mutase. Biochem. J. 337, 89–95 [PMC free article] [PubMed] [Google Scholar]
- 34. Engel M., Mazurek S., Eigenbrodt E., Welter C. (2004) Phosphoglycerate mutase-derived polypeptide inhibits glycolytic flux and induces cell growth arrest in tumor cell lines. J. Biol. Chem. 279, 35803–35812 [DOI] [PubMed] [Google Scholar]
- 35. Kanfi Y., Peshti V., Gozlan Y. M., Rathaus M., Gil R., Cohen H. Y. (2008) Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 582, 2417–2423 [DOI] [PubMed] [Google Scholar]
- 36. Imai S. (2009) SIRT1 and caloric restriction. An insight into possible trade-offs between robustness and frailty. Curr. Opin. Clin. Nutr. Metab. Care 12, 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imai S. (2009) Nicotinamide phosphoribosyltransferase (Nampt). A link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 15, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Imai S. (2009) The NAD World. A new systemic regulatory network for metabolism and aging–Sirt1, systemic NAD biosynthesis, and their importance. Cell Biochem. Biophys. 53, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases. Implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caton P. W., Nayuni N. K., Kieswich J., Khan N. Q., Yaqoob M. M., Corder R. (2010) Metformin suppresses hepatic gluconeogenesis through induction of SIRT1 and GCN5. J. Endocrinol. 205, 97–106 [DOI] [PubMed] [Google Scholar]
- 41. Gillum M. P., Erion D. M., Shulman G. I. (2011) Sirtuin-1 regulation of mammalian metabolism. Trends Mol. Med. 17, 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Caton P. W., Nayuni N. K., Khan N. Q., Wood E. G., Corder R. (2011) Fructose induces gluconeogenesis and lipogenesis through a SIRT1-dependent mechanism. J. Endocrinol. 208, 273–283 [DOI] [PubMed] [Google Scholar]
- 43. Wei D., Tao R., Zhang Y., White M. F., Dong X. C. (2011) Feedback regulation of hepatic gluconeogenesis through modulation of SHP/Nr0b2 gene expression by Sirt1 and FoxO1. Am. J. Physiol. Endocrinol. Metab. 300, E312–E320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.