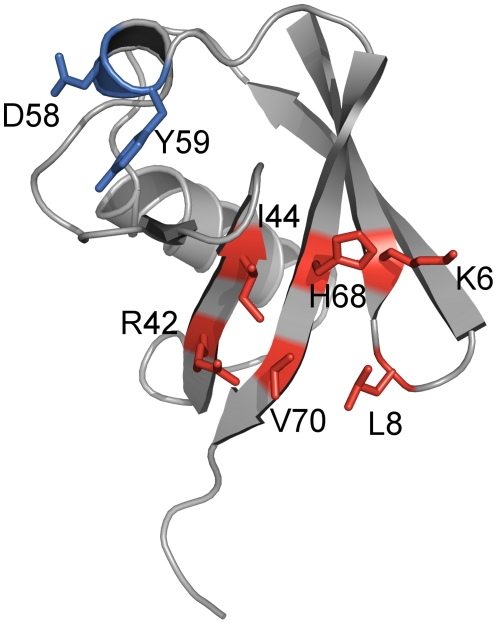
Figure 1. Amino acid positions chosen for randomization in library construction.
Based on the scaffold ubiquitin (shown here in cartoon representation), in particular with an F45W substitution, a library for the selection of artificial binding proteins was generated. For this purpose the six surface-exposed amino acid residues K6, L8, R42, I44, H68 and V70 (highlighted in red), located in the beta-sheet region of the scaffold, were chosen to be randomized for library construction. After in vitro selection against TNF-alpha, in the ubiquitin variant named 10F the residues D58 and Y59 (highlighted in blue) were found deleted. This figure was generated using pdb entry 1UBI and the software PyMOL version 0.99rc6 (DeLano Scientific LLC, South San Francisco, CA).