Figure 2. qPCR validation of RA target genes.
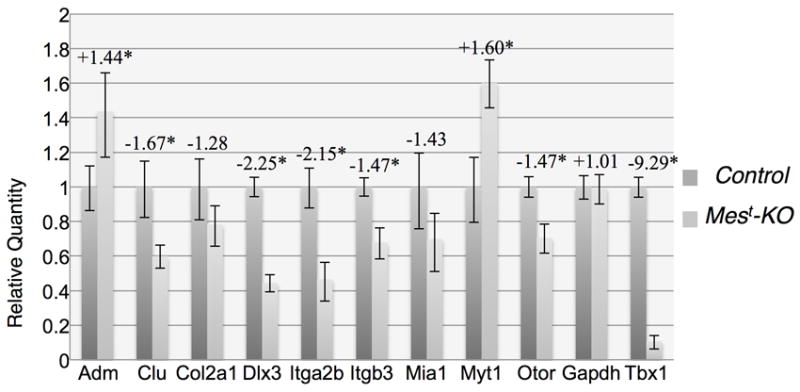
Genes shown to be regulated by RA in other systems (Table 1) were validated by qPCR on microdissected Mest-KO periotic regions (OV+POM) using TaqMan probes. As expected, Adm, Clu, Dlx3, Itga2b, Itgb3, Myt1, Otor, and control probe Tbx1 were significantly changed in T-Cre KO versus control ears. Col2a1, Mia1, and control probe Gapdh were not significantly changed, though both Col2a1 and Mia1 show a trend towards down-regulation that is not statistically significant. Fold change values are shown above (Mest-KO vs. Control) and asterisk indicates significance with p<0.05.
