Abstract
Background. A recent genome-wide association study reported a strong association with a single-nucleotide polymorphism (SNP) in the inosine triphosphate (ITPA) gene and hemolytic anemia in patients infected with hepatitis C virus (HCV) receiving pegylated interferon and ribavirin. We investigate these polymorphisms in a cohort of human immunodeficiency virus (HIV)/HCV–coinfected patients.
Methods. DNA was available for 161 patients with validated outcomes. We analyzed the association between the variants and week 4 hemoglobin reduction. Anemia over the course of therapy, ribavirin (RBV) dose reduction, serum RBV level, and rapid virological response (RVR) and sustained virological response (SVR) were also investigated. Using a candidate gene approach, ITPA variants rs1127354 and rs7270101 were tested using the ABI TaqMan kit. Multivariable models were used to identify predictors of anemia.
Results. A significant minority (33%) of patients were predicted to have reduced ITPase activity. The minor allele of each variant was associated with protection against week 4 anemia. In multivariable models only the genetic variants, creatinine, and zidovudine exposure remained significant. ITPase deficiency was not associated with RBV-dose reduction, RVR, or SVR.
Conclusions. This study confirms that polymorphisms in the ITPA gene are associated with protection from RBV-induced anemia in HIV/HCV-coinfected patients but not improved clinical outcomes.
Hepatitis C virus (HCV) chronically infects approximately 170 million people worldwide [1]. In resource-rich countries, HCV is the leading cause of end-stage liver disease and hepatocellular carcinoma, as well as the main indication of the need for liver transplantation [1]. In the United States and Europe, HCV affects 15%–30% of individuals infected with human immunodeficiency virus (HIV) and currently represents a major cause of morbidity and mortality in the coinfected population [2, 3]. Therapy for chronic hepatitis C with pegylated interferon alfa (pegIFN) plus ribavirin (RBV) is poorly tolerated, and only half of patients who receive therapy achieve a sustained virological response (SVR) [4]. This figure is lower in HIV/HCV-coinfected patients, who are at greater risk of hematologic adverse effects [5, 6]. Increased RBV dose has been shown to improve SVR rates in difficult to treat patient groups, and serum ribavirin levels have, in some studies, been shown to correlate with on-treatment response; however, higher ribavirin exposure may also increase adverse events, particularly RBV-induced hemolytic anemia [7–9].
A genomewide association study (GWAS) of a subset of HCV genotype-1 monoinfected patients from the IDEAL study recently reported a strong association between a single-nucleotide polymorphism (SNP) rs6051702 on chromosome 20 and treatment-related anemia in individuals with genotype 1 infection [10]. The responsible gene locus is in the inosine triphosphatase (ITPA) gene, which encodes inosine triphosphatase (ITPase), a protein that hydrolyses inosine triphosphase [10]. Previous reports of loss of function gene mutations have been described that lead to ITPase deficiency: a benign red cell enzymopathy characterized by accumulation of ITP in erythrocytes and increased toxicity of purine analog drugs [11, 12]. Genotyping of these variants demonstrated that the association signal was entirely explained by 2 functional variants in the ITPA gene: a missense variant in exon 2 (rs1127354) and a splice-altering SNP in intron 2 (rs7270101) [13–16]. The mechanism by which these mutations result in protection from treatment-induced anemia is in part related to the ability of ITP to substitute for erythrocyte guanosine triphosphate (GTP), which is depleted by RBV during biosynthesis of adenosine triphosphate (ATP) [17]. Recent data also suggest that ITPA variants may remain predictive in the setting of triple combination therapy with telaprevir plus pegIFN/RBV [18].
We sought to extend our understanding of this genetic association in an HIV/HCV-coinfected cohort of patients and in other HCV genotypes. Specifically, we investigate the association between the functional ITPA variants and the development of anemia at week 4, and the impact on hemoglobin levels over the course of therapy. We also explore whether these genetic variants are associated with serum RBV levels, RBV dose reductions, rapid virological response (RVR), and SVR.
METHODS
Study Population
Patients were recruited at Hospital Carlos III, a reference HIV clinic located in Madrid, Spain. From a cohort of 650 HIV/HCV-coinfected patients seen in the clinic between 2002 and 2008 with regular follow-up (characteristics described elsewhere [19]), we selected 198 interferon-naive individuals who had either completed a course of therapy with pegIFN/RBV and had validated outcomes or were discontinued on therapy based on standard stopping rules; thus, this is an adherent cohort. Written informed consent for the study of genetic material was obtained from all individuals, and the study protocol was evaluated and approved by the hospital ethics committee. Both plasma and peripheral blood mononuclear cells were stored on all patients.
All patients were treated with pegIFN alfa-2a or -2b at standard doses (180 μg/week or 1.5 μg/kg/week, respectively) plus weight-adjusted RBV (1000 mg/day for patients weighing <75 kg and 1200 mg/day for patients weighing >75 kg). Patients with HCV genotypes 1 or 4 received 48 or 72 weeks of treatment, and patients with HCV genotype 2 or 3 received 24 or 48 weeks of treatment. Early stopping rules were applied for subjects at weeks 12 and 24 [20]. All patients completed 24 weeks of follow-up after the end of treatment to assess for SVR. Dose adjustments for RBV were completed per clinic protocol. In brief, RBV dose was reduced by 200-mg/day decrements when hemoglobin fell below 10.0 g/dL, this was repeated on a weekly basis until hemoglobin either stabilized or decreased to <8.5 g/dL, at which time the RBV was discontinued. Use of growth factors was not permitted.
Genotyping
In total, 198 patients were genotyped at the polymorphic sites rs1127354 and rs7270101 on chromosome 20 using the ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems), as described elsewhere [10]. The possible genotypes for each biallelic polymorphism are rs1127354: C/C, A/C, A/A (minor allele = A); rs7270101: A/A, A/C, C/C (minor allele = C). The cohort was previously genotyped for the IL28B SNP rs12979860 associated with HCV treatment response [21, 22].
HCV Load and Genotyping
Plasma HCV-RNA was measured using a real-time polymerase chain reaction (PCR) assay (COBAS TaqMan, Roche), with a lower limit of detection of 10 IU/mL. HCV genotyping was performed using a commercial real-time (RT)-PCR hybridization assay (Versant HCV Genotype v2.0 LiPA, Siemens). Plasma HIV-RNA was measured using Versant HIV-1 RNA v3.0 (Siemens), with a lower limit of detection of 50 copies/mL. RBV plasma trough concentrations (RBV Ctrough) were variably measured as part of clinical care at weeks 4, 12, 24, 36, and 48. Early-morning samples were collected prior to the first daily dose of RBV (patient instructed to hold morning dose until labs drawn) and, on average, 12 hours after the last dose. Blood samples were centrifuged within 1 hour of sample collection. RBV Ctrough was quantified using a validated ultraviolet reverse-phase high-performance liquid chromatography [23].
Liver Fibrosis Staging
The extent of liver fibrosis was measured using transient elastography by FibroScan (Echosens) [24, 25]. The median value of all tests per patient is expressed in kilopascal (kPa) units. Advanced liver fibrosis (severe fibrosis or cirrhosis, corresponding to Metavir scores F3 and F4) was defined for liver stiffness values ≥9.5 kPa, according to previous reports from both HCV-monoinfected and HIV/HCV-coinfected patients [26, 27].
Definition of ITPase Deficiency Variable According to rs1127354/rs7270101 Genotypes
Severity of ITPase deficiency was defined according to previous studies [10], and a composite ITPase deficiency variable was created for analysis (Table 1). ITPase activity was defined by the accumulated presence of minor alleles at the respective polymorphic sites. As described previously, only 3 haplotypes were observed; thus the minor alleles for both SNPs never occur together on the same chromosome [10].
Table 1.
ITPase Deficiency Variable
| rs1127354 | rs7270101 | Predicted ITPase Activity, % | Predicted ITPase Deficiency |
| Wild type (C/C) | Wild type (A/A) | 100 | − |
| Wild type (C/C) | Heterozygosity (A/C) | 60 | + |
| Heterozygosity (C/A) | Wild type (A/A) | 30 | ++ |
| Wild type (C/C) | Homozygosity (C/C) | 30 | ++ |
| Heterozygosity (C/A) | Heterozygosity (A/C) | 10 | +++ |
| Homozygosity (A/A) | Wild type (A/A) | <5 | +++ |
Definition of an ITPase deficiency variable according to rs1127354/rs7270101 genotypes: severity of ITPase deficiency was predicated as absent, representing wild-type activity (−), mild (+), moderate (++), or severe (+++) deficiency, according to previously published studies [12].
Abbreviation: ITPase, inosine triphosphatase.
Study Objectives
For replication of the primary GWAS discovery, we analyzed hemoglobin reduction at week 4. The primary objectives included the association of ITPase deficiency with (1) absolute hemoglobin reduction as a continuous trait and (2) hemoglobin reduction >3 g/dL. Secondary objectives included the association of ITPase deficiency with (1) hemoglobin reduction over the course of therapy, defined both qualitatively and quantitatively (reduction >3 g/dL); (2) need for RBV dose reduction; (3) RBV serum level at week 4, defined both qualitatively and quantitatively (level <2 mg/L); (4) RVR; and (5) SVR.
Statistical Analysis
For descriptive statistics, continuous variables were summarized as median (lower and upper quartiles). Categorical variables were described as frequency and percentage. Comparisons between groups were performed using a Wilcoxon test for skewed continuous data or the χ2/Fisher exact test for categorical data. Significance was defined as P < .05 with a 2-tailed test. Association between the individual polymorphisms/ITPase deficiency variable and the anemia phenotypes were tested using single-marker genotype trend tests of association in linear or logistic regression models. Multivariable regression models with backward selection were used to identify independent predictors of anemia. A significance level of .05 was used for removal from the model. All statistical analyses were performed with SAS software, version 9.2 (SAS Institute).
RESULTS
Cohort Characteristics
The clinical characteristics of the study population are described in Table 2. In total, 161 patients were included in the final analysis due to lack of adequate DNA for genotyping of 37 patients. All patients were of European ancestry, and IL28B C/C genotype prevalence was 43%. Eighty-four percent of patients were receiving antiretroviral therapy (ART) (15% [24 of 161 patients] on zidovudine [AZT] and 29% [46 of 161 patients] on abacavir [ABC]), and 80% had undetectable plasma HIV-RNA. Median HCV RNA was 6.6 log10 IU/mL and the HCV genotype distribution was HCV-1, 59%; HCV-3, 29%; HCV-2, 1%; and HCV-4, 11%. Length of treatment was as follows: 24 weeks (24%, 39 of 161 patients), 48 weeks (50%, 81 of 161 patients), and 72 weeks (9%, 14 of 161 patients). Five patients (3%) did not have treatment duration available (although treatment end point was available), and on the basis of early stopping rules, 14% (22 of 161 patients) were discontinued on or before week 24. The overall SVR rate in this treatment-adherent cohort was 56% (90 of 161 patients).
Table 2.
Baseline Characteristics
| Characteristics | Cohort (N = 161) |
| Age, y, median (quartiles) | 42 (39, 46) |
| Male sex, No. (%) | 120 (75) |
| Median Hb level, g/dL | 15.6 (14.6, 16.5) |
| Median CD4 count, cells/μL | 500 (378, 698) |
| Patients on ART, No. (%) | 135 (84) |
| Median HIV RNA, log10 copies/mL | 1.7 (1.7, 1.8) |
| Patients with HIV suppression, <50 copies/mL | 125 (80) |
| On AZT, No. (%) | 24 (15) |
| On ABC, No. (%) | 46 (29) |
| RBV dose reduction, No. (%) | 18 (11) |
| Median HCV RNA, log10 IU/mL | 6.6 (4.8, 7.0) |
| HCV genotype, No. (%) | |
| 1/4 | 112 (70) |
| 2/3 | 49 (30) |
| Median liver stiffness, kPa | 6.2 (4.8, 9.6) |
Abbreviations: ABC, abacavir; ART, antiretroviral therapy; AZT, zidovudine; Hb, hemoglobin; HCV, hepatitis C virus; HIV, human immunodeficiency virus; RBV, ribavirin.
Population Distribution of ITPA Gene Variants
The cohort was genotyped for the 2 causal polymorphisms, rs1127354 and rs7270101. The minor allele (A) frequency for rs1127354 was 0.06, and the minor allele (C) frequency for rs7270101 was 0.12, with the population demonstrating Hardy-Weinberg equilibrium (P = .58 and P = .79, respectively). The population frequencies for both rs1127354 and rs7270101 are shown in Table 3. The majority of patients (65%) were predicted to have normal wild-type ITPase enzyme activity; however, a significant minority (33%) were predicted to have reduced ITPase activity (Table 3).
Table 3.
Genotype Frequency (rs1127354 and rs7270101) in the Study Population and Population Distribution of Predicted Level of ITPase Deficiency, According to ITPA Genotype
| Genotype | Population Frequency, No. (%) |
| rs1127354 | |
| CC | 140 (89) |
| CA | 18 (11) |
| AA | 0 |
| rs7270101 | |
| AA | 119 (75) |
| AC | 36 (23) |
| CC | 4 (2) |
| Predicted ITPase Deficiency | Population Distribution, No. (%) |
| − | 104 (67) |
| + | 32 (20) |
| ++ | 17 (11) |
| +++ | 3 (2) |
Abbreviations: ITPA, inosine triphosphate; ITPase, inosine triphosphatase.
ITPase Deficiency Protected Against Week 4 Anemia
The primary objective of the study included assessment of the pharmacogenomic association of ITPase deficiency with ribavirin-induced anemia at 4 weeks: hemoglobin reduction as a continuous trait and hemoglobin reduction >3 g/dL. Both ITPA variants individually and the composite ITPase deficiency variable were evaluated for an association with anemia (Table 4). Both ITPA variants and the ITPase deficiency variable were strongly associated with hemoglobin reduction at 4 weeks, with the minor allele having a protective effect in all cases (Table 4). The association of ITPase deficiency with week 4 anemia outcomes did not change with stratification by HCV genotype (data not shown). Compared to the individual alleles, the composite ITPase deficiency variable appeared to have the strongest association with protection against quantitative week 4 anemia, estimated to explain 23% of the variability of the hemoglobin reduction at week 4, similar to that reported by Fellay et al [10] (Table 4). There was no association between ITPase deficiency and baseline hemoglobin level (data not shown).
Table 4.
Regression Models for Quantitative Hemoglobin Reduction at Week 4 and Hemoglobin Reduction >3 g/dL at Week 4
| ITPA Variants and Week 4 Quantitative Hb Reductiona | ||||||
| ITPA Variant | Estimate | Standard Error | P Value | Adjusted Estimate | Standard Error | Adjusted P Valueb |
| rs1127354 | −1.85 | 0.42 | <.0001 | −1.75 | 0.56 | .003 |
| rs7270101 | −0.87 | 0.25 | .0008 | −1.20 | 0.40 | .004 |
| Composite ITPase Deficiency Variable and Week 4 Quantitative Hb Reductionc | ||||||
| Parameter | Estimate | Standard Error | P Value | |||
| ITPase deficiency | −0.97 | 0.25 | .0003 | |||
| Creatinine | −1.62 | 0.78 | .043 | |||
| Zidovudine exposure | 1.79 | 0.45 | .0002 | |||
| ITPA Variants and Week 4 Hb Reduction >3 g/dLa | ||||||
| ITPA Variant | OR | 95% CI | P Value | Adjusted OR | 95% CI | Adjusted P Valueb |
| rs1127354 | 0.10 | .01–.82 | .03 | 0.12 | .01–1.38 | .089 |
| rs7270101 | 0.24 | .09–.63 | .004 | 0.21 | .05–.92 | .038 |
| Composite ITPase Deficiency Variable and Week 4 Hb Reduction >3 g/dLc | ||||||
| Parameter | OR | 95% CI | P Value | |||
| ITPase deficiency | 0.24 | .08–.73 | .012 | |||
| Creatinine | 0.04 | .002–.93 | .049 | |||
| Zidovudine exposure | 14.9 | 1.6–135 | .016 | |||
Abbreviations: CI, confidence interval; Hb, hemoglobin; ITPA, inosine triphosphate; ITPase, inosine triphosphatase; OR, odds ratio.
Covariables: exposure to zidovudine and baseline creatinine. Sex, age, baseline CD4 count, liver stiffness, baseline hemoglobin level, week 4 ribavirin level, and ribavirin dose by weight were removed by backward selection.
Adjusted estimates, P values were calculated in models in which the other functional variant was already included.
Sex, age, baseline CD4 count, liver stiffness, baseline hemoglobin level, week 4 ribavirin level, and ribavirin dose by weight were removed by backward selection.
In the multivariable analyses, the ITPase deficiency variable and allele variants maintained a strong association with protection from week 4 anemia and severity of week 4 anemia (Table 4). Other variables associated with risk or severity of week 4 anemia included higher baseline creatinine and exposure to AZT. Variables not associated with week 4 anemia included age, sex, baseline HIV load, baseline CD4 cell count, baseline hemoglobin level, weight, liver stiffness, exposure to ABC, IL28B genotype, RBV dose, and RBV dose change.
Predictors Associated With Anemia in Those Patients Not Exposed to AZT
AZT is known to cause macrocytic anemia and thus is a confounder in this analysis. To account for confounding, the association of ITPase deficiency and anemia was analyzed in 2 ways: in a multivariable analysis including AZT with an interaction term as a predictor variable reported above and in a multivariable analysis stratified by AZT exposure. In the analysis excluding AZT, both ITPA variants and the ITPase deficiency variable remained strongly associated with hemoglobin reduction at 4 weeks (quantitative and qualitative) (data not shown). When included in multivariable models; in addition to the ITPase deficiency variable, hemoglobin baseline, week 4 RBV level, and RBV dose by weight remained positively associated with week 4 anemia.
Predictors Associated With Anemia Throughout the Course of HCV Therapy
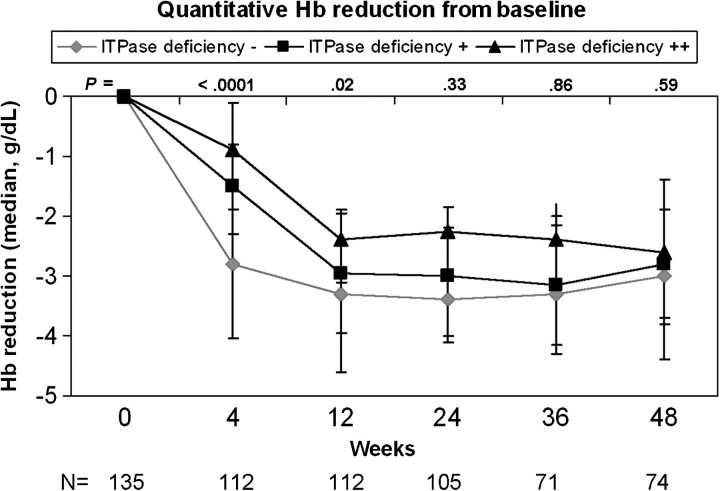
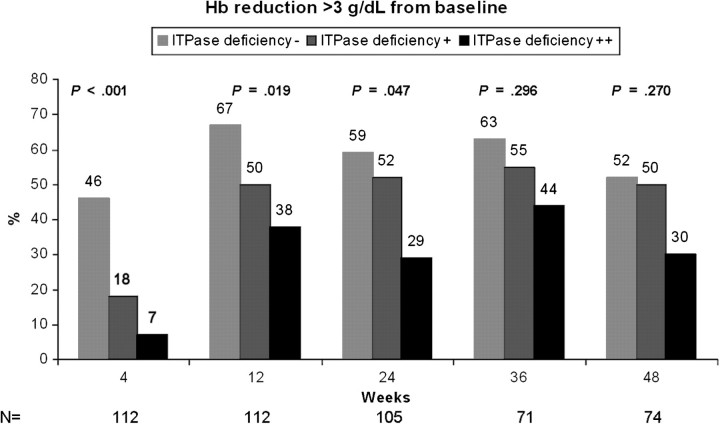
The association with the ITPase deficiency variable and anemia was evaluated throughout the course of therapy for the majority of patients not exposed to AZT (n = 137). ITPase deficiency was associated with a delayed hemoglobin decline in the first 12 weeks of therapy, and this resulted in less absolute hemoglobin reduction over the course of therapy (Figure 1). In those patients with moderate ITPA deficiency, only 7% of patients experienced hemoglobin reduction >3 g/dL at week 4 of therapy, as compared with 46% of those patients with wild-type ITPase activity (P = .001, Figure 2). The frequency of clinically significant anemia increased for all groups over the course of therapy, with 59%, 52%, and 29% of patients with no, moderate, or high ITPase deficiency, respectively, achieving hemoglobin reduction >3 g/dL at week 24 (P = .03, Figure 2). Differences at later time points did not reach statistical significance. Results were similar when stratified by HCV genotype (data not shown). Stratification of the model by need for RBV dose reduction did not change the association of the ITPase deficiency variable and anemia throughout the course of therapy (data not shown).
Figure 1.
ITPase deficiency variable was associated with less reduction in median hemoglobin (Hb) level at all time points, although this did not meet statistical significance at later weeks.
Figure 2.
ITPase deficiency variable was associated with less absolute reduction in median hemoglobin (Hb) level at all time points, although this did not meet statistical significance at later weeks.
ITPase Deficiency Is Associated With Lower Serum Ribavirin Levels But Not With Ribavirin Dose Reduction
RBV dose reduction was uncommon in this adherent Spanish cohort, with only 18 patients (11%) requiring dose reduction during therapy. Those patients who required dose reduction were less likely to achieve SVR (P = .025). When the relationship between the ITPA gene variants (ITPase variable, rs1127354, or rs7270101) and dose reduction was examined, no relationship was noted (P = .69, P = .97, and P = .07, respectively), although a trend was evident for rs7270101.
Serum RBV trough levels were available on 60% of patients at weeks 4 and 12, and 25%–44% at weeks 24, 36, and 48. At week 4, serum ribavirin troughs were different across ITPase deficiency levels (−deficiency median, 2.58 mg/dL [quartiles; 1.71, 3.01]; +deficiency, 1.91 mg/dL [1.65, 2.70]; ++deficiency, 1.65 mg/dL [1.44, 2.46]; P = .03) but appeared to converge between weeks 4 and 12, with no difference between deficiency levels by week 12 (median, 2.5 mg/dL [quartiles; 1.54, 2.88], P = .86). ITPase deficiency was associated with lower serum RBV levels at week 4 of therapy, when modeled as a qualitative or quantitative (<2 mg/dL) univariable (P = .026 and P = .019, respectively). In the univariable analysis, no other variable was significantly associated with week 4 RBV level including weight, RBV dose, or creatinine. However, when these variables were included in a multivariable regression, the association with week 4 RBV level and ITPase deficiency was no longer significant.
ITPase Deficiency Is Not Associated With RVR or SVR
ITPase deficiency was tested for an association with SVR in a logistic regression model that included age, sex, baseline HCV RNA (<600 000), HCV genotype (1/4 vs 2/3), RBV dose change, IL28B genotype, and liver stiffness (Table 5). No association with SVR was found for ITPase deficiency. Results were similar when stratified by HCV genotype (data not shown). No association was noted with ITPase deficiency and RVR; however, hemoglobin reduction >3 g/dL at 4 weeks and lower week 4 hemoglobin level were associated with higher rates of RVR. This finding did not translate to differences in SVR rates (data not shown).
Table 5.
Composite ITPase Deficiency Variable Is Not Associated with Sustained Virological Response
| Parameter | OR | 95% CI | P Value |
| ITPase deficiency variable | 1.65 | 0.71–3.86 | .245 |
| Age | 0.87 | 0.76–1.00 | .059 |
| Female sex | 0.84 | 0.16–4.47 | .843 |
| Liver stiffness | 0.83 | 0.72–0.97 | .017 |
| IL28B TT or CT | 0.08 | 0.02–0.33 | .0006 |
| Baseline HCV RNA <600,000 | 30.3 | 4.74–193 | .0003 |
| No RBV dose reduction | 0.21 | 0.03–1.43 | .111 |
Abbreviations: CI, confidence interval; HCV, hepatitis C virus; ITPase, inosine triphosphatase; OR, odds ratio; RBV, ribavirin.
DISCUSSION
In this study we evaluated the role of the recently discovered ITPA genetic variants that protect against RBV-induced hemolytic anemia, in an HIV/HCV-coinfected cohort. This issue is of great clinical importance for HIV-coinfected individuals, who are documented to have higher rates of anemia on treatment. To our knowledge, this is the first study to examine the ITPA genetic variants in an HIV-coinfected population and to explore the association of ITPase deficiency and serum RBV levels.
One-third (33%) of the cohort was predicted to have reduced ITPase activity and was found to be relatively protected from the development of anemia early in the treatment course. ITPase deficiency was a strong predictor of protection from week 4 anemia, including in those patients on concomitant AZT. Patients with at least moderate deficiency appeared to receive the most protection, with maximal benefit early in therapy, and persistence throughout the course of therapy. The rate of hemoglobin decline was also more rapid in those patients with wild-type ITPase activity, and this correlated to greater absolute decreases in hemoglobin level over the course of therapy. These results were the same regardless of HCV genotype. These data are in agreement with recent studies in HCV-monoinfected patients [28, 29].
Although those patients with reduced ITPase activity were protected from developing severe anemia throughout the course of therapy, this did not translate into differences in important clinical outcomes such as need for RBV dose reduction, RVR, or SVR. Thompson et al [28, 29] reported that ITPase deficiency was associated with less RBV dose reduction in HCV genotype 1 patients for whom RBV dose was reduced due to anemia, but not in HCV genotype 2/3 patients. Dose reduction was also reported more frequently in those without reduced ITPase activity in the first 12 weeks of triple combination therapy with telaprevir plus pegIFN/RBV [18]. There are 2 likely reasons for our inability to show a difference in RBV dose: (1) our treatment-adherent cohort infrequently required dose reduction (11% vs 46%), and (2) our cohort was heterogeneous, including all HCV genotypes. No study to date has found that ITPA deficiency and the conferred protection against RBV-induced anemia are associated with treatment response outcomes [18, 28, 29]. The reason for this is unknown, given other data suggesting that severity of week 4 anemia is associated with SVR [30]. However, given the low frequency of the minor ITPA alleles in the study population, it is possible that these studies, including ours, were not adequately powered to observe an effect of ITPA variants on treatment outcomes. Furthermore, due to confounding of rate of decline of hemoglobin and dose reduction on treatment outcomes, we completed stratified analyses by using these variables but were again limited by power and unable to show an association with treatment outcomes.
One well-recognized etiology of anemia in the HIV/HCV-coinfected patient receiving HCV therapy is concomitant administration of AZT, a thymidine analog. The mechanism of AZT-induced anemia is not well understood. One study reported that AZT decreased globin gene expression, resulting in altered development and differentiation of erythroid cells [31]. Here, exposure to AZT was the strongest predictor of week 4 anemia and of anemia throughout the course of therapy. ITPase deficiency provided significant protection from RBV-induced anemia in the setting of concomitant AZT use; however, its protective effect over the course of therapy was obscured in those patients on AZT. This may be due to the inability of patients exposed to both RBV and AZT to have an appropriate bone marrow response to the RBV-induced hemolytic anemia, in the setting of AZT-associated bone marrow suppression. Although AZT is not recommended for concomitant administration with ribavirin and was associated with the development of anemia, it was not associated with more dose reduction or treatment outcomes, which suggests that it may be used if other options are not feasible.
The relationship between ITPA variants and anemia is complex, and the effect of the ITPA variants on ribavirin levels and pharmacodynamics has not to our knowledge been explored previously. Although limited because of the size and scope of the study, in a univariable analysis ITPase deficiency was associated with lower serum RBV levels at week 4 of therapy. This association was no longer significant when built into a multivariable analysis that included baseline creatinine, RBV dose, and weight. How serum RBV levels correlate to intraerythrocytic RBV-triphosphate—the metabolite within the red blood cell that is thought to lead to oxidative damage to the red cell membrane and hence intravascular destruction—is unknown [32]. Due to the explorative nature of these findings, additional prospective study of the impact of ITPA deficiency on serum and intracellular RBV levels is necessary to validate this association.
There are several limitations of this study that deserve mention. In an attempt to compare the most extreme outcome phenotypes, only those patients with treatment adherence (discontinuation due to defined treatment stopping points) were included in the study. This may have resulted in the exclusion of patients who were intolerant of RBV and required both dose reduction and treatment discontinuation, thus underestimating the number of these events occurring in this population. Furthermore, because of the retrospective nature of the study design, there was loss of data points (hemoglobin and RBV level) at later treatment dates, introducing risk for type II error, and the availability of clinically important symptoms that are associated with anemia (such as fatigue and shortness of breath) were not available to include in the analysis.
In conclusion, functional variants of the ITPA gene, associated with ITPase deficiency, are protective against RBV-induced hemolytic anemia in HIV/HCV-coinfected patients. Although this difference was greatest in the first 4 weeks, it did persist throughout the treatment course. This protection is similar among different HCV genotypes. Although patients with ITPase deficiency had less anemia, in this study that benefit did not correlate to less dose reduction or clinically relevant end points such as RVR and SVR. For now, the use of assessing the ITPA genotype in a patient prior to initiating therapy with RBV is unclear. In the HIV/HCV-coinfected population, symptoms of severe anemia, including shortness of breath and fatigue, can be limiting and lead to discontinuation of therapy. Thus, potential roles for the use of pretreatment screening could include identifying patients who are more likely to tolerate higher doses of RBV or who are at greater risk of more severe anemia. In the real world setting, such knowledge could impact clinic outcomes, including need for growth factor use and dropout due to intolerance of therapy, and is likely to remain relevant in the setting of first-generation HCV protease inhibitors.
Notes
Author contributions.
Study design: S. N., N. R., A. T., P. C., J. G. M., V. S.; data collection: N. R., J. B., J. M., S. R., E. V.; data analysis: S. N.; laboratory analysis: K. S., D. G.; access to data, manuscript writing, and review: all authors.
Acknowledgments.
The authors thank all patients who participated in the study.
Financial support.
This work was supported by the National Institutes of Allergy and Infectious Diseases, 1K23-A1096913-01 (S.N.).
Potential conflicts of interest.
J. G. M., A. T., and D. G. are coinventors of a patent application based on the ITPA discovery. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2005;29:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Graham C, Baden L, Yu E, et al. Influence of HIV infection on the course of hepatitis C virus infection: a meta-analysis. Clin Infect Dis. 2001;33:562–9. doi: 10.1086/321909. [DOI] [PubMed] [Google Scholar]
- 3.Weber R, Sabin C, Friis-Møller N, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166:1632–41. doi: 10.1001/archinte.166.15.1632. [DOI] [PubMed] [Google Scholar]
- 4.Ghany MG, Strader DB, Thomas DL, Seeff LB. AASLD practice guidelines: diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–74. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung RT, Anderson J, Volberding P, et al. Peginterferon alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–9. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torriani FJ, Rodrguez-Torres M, Rockstroh JK, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–50. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 7.Nunez M, Miralles C, Berdun MA, et al. Role of weight-based ribavirin dosing and extended duration of therapy in chronic hepatitis C in HIV-infected patients: the PRESCO trial. AIDS Res Hum Retroviruses. 2007;23:972–82. doi: 10.1089/aid.2007.0011. [DOI] [PubMed] [Google Scholar]
- 8.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1060–9. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 9.Maynard M, Gagnieu MC, Pradat P, Bailly F, Trepo C. Relevance of ribavirin plasma concentration for the prediction of treatment response. J Hepatol. 2005;42(Suppl 2):224. [Google Scholar]
- 10.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–8. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 11.Bierau J, Lindhout M, Bakker JA. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics. 2007;8:1221–8. doi: 10.2217/14622416.8.9.1221. [DOI] [PubMed] [Google Scholar]
- 12.Stocco G. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85:164–72. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumi S. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–7. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 14.Cao H, Hegele RA. DNA polymorphisms in ITPA including basis of inosine triphosphase deficiency. J Hum Genet. 2002;47:620–2. doi: 10.1007/s100380200095. [DOI] [PubMed] [Google Scholar]
- 15.Arenas M, Duley J, Sumi S, Sanderson J, Marinaki A. The ITPA c.94C>A and g.IVS2+21A>C sequence variants contribute to missplicing of the ITPA gene. Biochem Biophys Acta. 2007;1772:96–102. doi: 10.1016/j.bbadis.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Shipkova M, Lorenz K, Oellerich M, Wieland E, von Ahsen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a caucasian population. Clin Chem. 2006;52:240–7. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 17.Hitomi Y, Cirulli ET, Fellay J, et al. Inosine triphosphate protects against ribavirin-induced ATP loss by restoring adenylosuccinate synthase function. Gastroenterology. 2011;140:1314–21. doi: 10.1053/j.gastro.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki F, Suzuki Y, Akuta N, et al. Influence of ITPA polymorphisms on decreases of hemoglobin during treatment with pegylated interferon, ribavirin, and telaprevir. Hepatology. 2011;53:415–21. doi: 10.1002/hep.24058. [DOI] [PubMed] [Google Scholar]
- 19.Medrano J, Resino S, Vispo E, et al. Hepatitis C virus (HCV) treatment uptake and changes in the prevalence of HCV genotypes in HIV-HCV coinfected patients. J Viral Hep. 2011;18:325–30. doi: 10.1111/j.1365-2893.2010.01309.x. [DOI] [PubMed] [Google Scholar]
- 20.Soriano V, Puoti M, Sulkowski M, et al. Care of patients coinfected with HIV and hepatitis C virus: 2007 updated recommendations from the HCV-HIV International Panel. AIDS. 2007;21:1073–89. doi: 10.1097/QAD.0b013e3281084e4d. [DOI] [PubMed] [Google Scholar]
- 21.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 22.Rallon NI, Naggie S, Benito JM, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–9. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morello J, Rodriguez-Novoa S, Cantillano AL, et al. Measurement of ribavirin plasma concentrations by high-performance liquid chromatography using a novel solid-phase extraction method in patients treated for chronic hepatitis. Ther Drug Monit. 2007;29:802–6. doi: 10.1097/FTD.0b013e31815bddf3. [DOI] [PubMed] [Google Scholar]
- 24.Ziol M, Handra-Luca A, Kettaneh A, et al. Non-invasive assessment of liver fibrosis by measurement of stiffness in patients with chronic hepatitis C. Hepatology. 2005;41:48–54. doi: 10.1002/hep.20506. [DOI] [PubMed] [Google Scholar]
- 25.Castera L, Vergniol J, Foucher J, et al. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. 2005;128:343–50. doi: 10.1053/j.gastro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 26.de Ledinghen V, Douvin C, Kettaneh A, et al. Diagnosis of hepatic fibrosis and cirrhosis by transient elastography in HIV/hepatitis C virus-coinfected patients. J Acquir Immune Defic Syndr. 2006;41:175–9. doi: 10.1097/01.qai.0000194238.15831.c7. [DOI] [PubMed] [Google Scholar]
- 27.Kirk G, Astemborski J, Mehta S, et al. Assessment of liver fibrosis by transient elastography in persons with hepatitis C virus infection or HIV-hepatitis C virus coinfection. Clin Infect Dis. 2009;48:963–72. doi: 10.1086/597350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson AJ, Fellay J, Patel K, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–9. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson AJ, Santon R, Piazzollo V, et al. ITPA genetic variants are protective against anemia during antiviral therapy for G 2/3 HCV, but do not decrease the need for RBV dose reduction or increase SVR. Hepatology. 2010;52:785A. doi: 10.1002/hep.24068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulkowski MS, Shiffman ML, Afdhal NH, et al. Hepatitis C virus treatment-related anemia is associated with higher sustained virologic response rate. Gastroenterology. 2010;139:1602–11. doi: 10.1053/j.gastro.2010.07.059. [DOI] [PubMed] [Google Scholar]
- 31.Spiga MG, Weidner DA, Trentesaux C, LeBoeuf RD, Sommadossi JP. Inhibition of beta-globin gene expression by 3′-azido-3′-deoxythymidine in human erythroid progenitor cells. Antivir Res. 1999;44:167–77. doi: 10.1016/s0166-3542(99)00065-0. [DOI] [PubMed] [Google Scholar]
- 32.De Franceschi L, Fattovich G, Turrini F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]