Abstract
Pig models of cystic fibrosis (CF) have recently been established that are expected to mimic the human disease closer than mouse models do. The human CLCA (originally named chloride channels, calcium-activated) member hCLCA4 is considered a potential modifier of disease severity in CF, but its murine ortholog, mCLCA6, is not expressed in the mouse lung. Here, we have characterized the genomic structure, protein processing, and tissue expression patterns of the porcine ortholog to hCLCA4, pCLCA4a. The genomic structure and cellular protein processing of pCLCA4a were found to closely mirror those of hCLCA4 and mCLCA6. Similar to human lung, pCLCA4a mRNA was strongly expressed in porcine lungs, and the pCLCA4a protein was immunohistochemically detected on the apical membranes of tracheal and bronchial epithelial cells. This stands in sharp contrast to mouse mCLCA6, which has been detected exclusively in intestinal epithelia but not the murine lung. The results may add to the understanding of species-specific differences in the CF phenotype and support the notion that the CF pig model may be more suitable than murine models to study the role of hCLCA4.
Keywords: CLCA, porcine, animal model, cystic fibrosis pig
The complex mechanisms of disease in cystic fibrosis (CF, mucoviscidosis) are still poorly understood, and specific therapies that target the basic genetic defect are currently out of reach. With the recent generation of porcine CF models (Rogers, Stoltz, et al. 2008), these aims may be achieved more easily because CF pigs develop all hallmark features of human CF, including developmental changes in the respiratory tract and lung disease (Meyerholz, Stoltz, Namati et al. 2010; Meyerholz, Stoltz, Pezzulo et al. 2010; Stoltz et al. 2010). Thus, the porcine CF model may have numerous advantages when compared to the murine models, which are far less than optimal to mimic the complex human CF pathology (Scholte et al. 2004).
CLCA proteins, originally named chloride channels, calcium-activated, mediate a constitutively expressed endogenous chloride current when heterologously expressed in different cell lines (Loewen et al. 2002; Hamann et al. 2009). Some of these proteins, including the human hCLCA2, are anchored to the cell membrane (Elble et al. 2006), whereas others are secreted, non-transmembrane proteins, including the human hCLCA1, the murine mCLCA3 (Gibson et al. 2005; Mundhenk et al. 2006), and the porcine pCLCA1 (Plog et al. 2009). A modulatory role has been discussed for several CLCA proteins in human CF and murine models of CF. The human hCLCA1/hCLCA4 gene locus acts as a modifier of residual anion conductance in rectum biopsies of CF patients (Ritzka et al. 2004), and a hCLCA1 allelic variant is associated with the severity of the intestinal CF phenotype in humans (van der Doef et al. 2010). A modulatory role has also been demonstrated for their murine orthologs. Experimental overexpression of the murine ortholog to hCLCA1, mCLCA3, ameliorates the intestinal pathology of CF mice (Young et al. 2007). Furthermore, mCLCA6, the murine ortholog to hCLCA4, is coexpressed with the murine CFTR protein in non–goblet cell enterocytes and possibly interacts with CFTR-associated pathways in this cell type (Bothe et al. 2008).
Several CLCA members have been discovered in different species, including human, mouse, pig, rat, cow, horse, and dog (Cunningham et al. 1995; Gandhi et al. 1998; Gruber, Elble, et al. 1998; Gruber, Gandhi, et al. 1998; Gaspar et al. 2000; Loewen et al. 2002; Anton et al. 2005; Jeong et al. 2005; Loewen and Forsyth 2005). Although the family is highly conserved in mammals, there are significant species-specific differences in the genomic structures and tissue expression patterns of direct CLCA orthologs. For example, the human gene cluster consists of four members, whereas there are eight murine CLCA genes instead (Patel et al. 2009). Recently, we reported that the porcine CLCA gene locus consists of five distinct CLCA genes (Plog et al. 2009). As an additional example for species-to-species variation, the human hCLCA1, the murine mCLCA3, and the porcine pCLCA1 are direct orthologs belonging to one phylogenetic cluster and are expressed in mucus-producing cells (Gruber, Elble, et al. 1998; Gruber, Gandhi, et al. 1998; Agnel et al. 1999; Leverkoehne and Gruber 2002; Plog et al. 2009). However, there are major differences in their tissue and cellular expression patterns. Neither hCLCA1 nor mCLCA3 has been found in the pancreatic or salivary ducts, the gall bladder, or the common bile duct despite strong expression of pCLCA1 in the respective porcine tissues (Gruber, Elble, et al. 1998; Leverkoehne and Gruber 2002; Loewen et al. 2005; Plog et al. 2009). In contrast, pCLCA1 has not been detected in the genital tract, whereas the mCLCA3 protein was found in uterine goblet cells and hCLCA1 RNA was detected in the uterus (Agnel et al. 1999; Leverkoehne and Gruber 2002). Interspecies variance is also obvious when comparing the expression patterns of the murine mCLCA5 with that of its human counterpart hCLCA2. The latter was localized to the basement membranes of basal epithelial cells, whereas mCLCA5 was detectable in keratohyalin granules of squamous epithelia only (Connon et al. 2004; Braun et al. 2010).
Species-specific differences in the anatomy, physiology, and expression patterns of relevant proteins are of prime interest for characterizing and interpreting animal models. For example, in a direct comparison of human, murine, and porcine CFTR proteins, Ostedgaard et al. (2007) identified important differences among the three species in the processing of this protein. Several mouse models have been intensely characterized, and the murine phenotype of CF is overall dominated by the intestinal disease. Neither lung disease nor fibrotic lesions of the pancreas typically seen in human CF patients are present in murine models (Davidson and Rolfe 2001; Scholte et al. 2004). Furthermore, interspecies variations have been observed in several other models of human diseases, including murine models for the characterization of secretory leukocyte protease inhibitors (Zitnik et al. 1997) and in studies on osteogenesis and tissue engineering of bone from murine cells (Pereira et al. 2009). In recent years, pigs have emerged as promising models for human diseases, due to the excellent comparability of their anatomy and the biochemical and genomic organization largely shared between humans and pigs (Rogers, Abraham, et al. 2008).
The porcine CF model may also become important to identify and study modifier genes, particularly those that may play a role in modulating pulmonary CF. Knowledge on their expression patterns in the pig and interspecies variances will be indispensable for interpreting CF in the pig. The aim of this study was therefore to characterize the genomic organization and protein processing, as well as mRNA and protein expression patterns, of pCLCA4a—the porcine ortholog to the putative human CF modifier hCLCA4 (Ritzka et al. 2004). We then compared key characteristics potentially relevant for CF with the human and murine orthologs hCLCA4 and mCLCA6, respectively.
Material and Methods
Bioinformatic Characterization of the pCLCA4a Genomic Structure in Comparison to hCLCA4 and mCLCA6
Exon–intron structures of the human, murine, and porcine CLCA orthologs were compared as described previously (Plog et al. 2009). In brief, recently identified sequences from porcine bacterial artifical chromosomes CH242-148M17 and CH242-483E7 were aligned with the mRNA sequence of the human hCLCA4 by BLAST search (http://blast.ncbi.nlm.nih.gov/blast.cgi). Exon structures of the pCLCA4a were identified by their analogy to the described structure of hCLCA4 (GenBank accession no. NM_012128.3) and to the structure of mClca6 (GenBank accession no. NW_001030737) as well as the structure of the splice donor and acceptor sites in the porcine bacterial artifical chromosome sequences.
Animals and Tissue Processing
Tissues from three male 6-week-old pigs, EUROC × Pietrain, and genital tracts of two additional female pigs, 2- and 3-month-old mixed breeds, that had been euthanized for other reasons and used in recent studies (Plog et al. 2009; Plog et al. 2010) were included in this study. All experiments were approved according to the rules of the German Animal Welfare Law by the Landesamt für Gesundheit und Soziales, Berlin (registration no. G 0323/06).
The following tissues were immersion fixed in 4% neutral buffered formaldehyde or shock frozen in liquid nitrogen after brief immersion in 2-methylbutane: nasal cavity, larynx, trachea, cranial left lung lobe, left main lung lobe, accessory lung lobe, tracheal bronchus, left principal bronchus, oesophagus, glandular and nonglandular stomach, duodenum, jejunum, ileum, caecum, colon, rectum, parotid salivary gland, pancreas, liver, gall bladder, kidney, urinary bladder, spleen, heart, aorta, brain cortex, cerebellum, medulla, eyes, skin from the perineum, rooting disc and prepuce, testicles, epidymides, spermatic cord, uterus, and ovaries.
Cloning and Sequencing of pCLCA4a
Total RNA was extracted from a porcine jejunal sample using the Trizol method (Invitrogen, Karlsruhe, Germany) and purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, including a digestion with DNase I (Qiagen). In sum, 750 ng of total RNA were reverse transcribed (IScript; Bio-Rad Laboratories, Inc, Hercules, CA) at 25C for 5 min, 42C for 30 min, and 85C for 5 min. PCR primers were designed (GenBank accession nos. CU694822 and CU469041) to amplify the open reading frame of pCLCA4a (upstream 5′-GAAAAGCCTCAACAAG-3′; downstream 5′-ATTTKATGAATGATC-3′). High Fidelity Polymerase (Fermentas, St Leon-Rot, Germany) was used for PCR amplification (1.5 units per 25 µl of reaction). PCR conditions were 10 cycles of 94C for 3 min, 94C for 30 sec, 44C for 30 sec, and 68C for 3 min 20 sec and 25 cycles of 94C for 30 sec, 44C for 30 sec, 68C for 3 min 20 sec, with a time increment of 10 sec per cycle and a final extension at 68C for 10 min. PCR products were controlled for specificity by endonuclease digestion with restriction enzymes EcoRV and HindIII (Fermentas) according to the manufacturer’s protocol.
T-addition to the EcoRV-digested and linearized pcDNA3.1 expression vector and A-addition to the pCLCA4a open reading frame were performed using GoTaq Flexi DNA Polymerase (Promega, Mannheim, Germany) and 10 mM dTTP or dATP, respectively (Invitrogen) in the presence of 25 mM MgCl2 (Promega, Mannheim, Germany) for 30 min at 70C. Ligation was performed using T4 DNA Ligase (Fermentas) at 16C overnight. The clone was completely sequenced and confirmed to represent the pCLCA4a ORF.
Computer-Aided Sequence Analyses and Generation of Antibodies
The pCLCA4a amino acid sequence was analyzed using the SignalP 3.0 (Nielson et al. 1997), Kyte–Doolittle (Kyte and Doolittle 1982), SOSUI (Hirokawa et al. 1998), HMMtop (Tusnady and Simon 2001), DAS (Cserzo et al. 1997), and PSORT II (Nakai and Horton 1999) software. Potential N-linked glycosylation sites were identified using the software NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc). Two oligopeptides were synthesized per immunogenicity prediction, one located in the amino-terminal cleavage product of pCLCA4a (p4a-N-1, corresponding to aa 355–368, DLIQITGSNERDKL) and one located in the predicted carboxy-terminal cleavage product (p4a-C-1, corresponding to aa 866–879, EADYTPDDSHPDPG). Oligopeptides were coupled to keyhole limpet hemocyanin and used for standard immunization of two rabbits each. Preimmune sera were collected before immunization and used as control in the immunodetection experiments. The four antisera obtained were designated p4a-N-1a, p4a-N-1b, p4a-C-1a, and p4a-C-1b.
Immunoblot Analyses and Immunoprecipitation
HEK293 cells were transiently transfected as described previously (Plog et al. 2009). Twenty-four hr after transfection, the cells were incubated in DMEM supplemented with 2% glutamate for 5 hr. Medium was removed, and cells were lysed on ice in 100 µl of standard lysis buffer (25 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.5% DOC, 0.5% Triton X-100) and supplemented with protease inhibitor tablets (Roche, Mannheim, Germany). To remove cellular fragments, medium samples were spun at 2,400 × g for 5 min at 4C, the pellet discarded, and the cell culture supernatant spun at 13,000 × g for 15 min at 4C. The second supernatant was ethanol precipitated overnight at −20C, spun at 14,000 × g for 20 min at 4C, and pellets were resuspended in 100 µl of standard lysis buffer. All tissue samples were homogenized in 1 ml of standard lysis buffer as described above using a Precellys 24 homogenizer (peqlab Biotechnologie GmbH, Erlangen, Germany). Forty µg of tissue lysates were subjected to immunoblot analyses.
Medium and lysate samples were boiled in standard 3-fold SDS-PAGE Laemmli buffer and separated by SDS-PAGE. Subsequent electroblotting onto PVDF membranes was followed by blocking the membranes in 1× Roti-Block (Carl Roth, Karlsruhe, Germany) for 1 hr at room temperature. The membranes were probed at 4C over night with anti-pCLCA4a antibodies p4a-N-1b or p1-C-1a, the preimmune sera, or the specific antibodies preincubated with their respective peptides as described previously (Plog et al. 2010). Briefly, antibodies were preabsorbed using 20 µg/ml of the respective specific peptides or an irrelevant peptide for 1 hr at room temperature under vigorous shaking. Specific and control antibodies were diluted 1:5000. Membranes were incubated with secondary horseradish peroxidase–conjugated anti-rabbit IgG for 1 hr, and protein labeling was visualized using enhanced chemiluminescence (Thermo Fisher Scientific, Rockford, IL). In a second step, membranes were stripped using stripping buffer (1.5% w/v glycine, 0.1% w/v SDS, 1.0 v/v Tween 20, pH 2.2) for 30 min at room temperature and subsequently probed overnight with anti-β-actin antibody (Sigma-Aldrich, Munich, Germany) diluted 1:100,000 at 4C.
pCLCA4a-transfected HEK293 cell lysates and medium were immunoprecipitated as described (Mundhenk et al. 2006). In brief, after preclearing of medium and cell lysates with protein A-Sepharose beads (Sigma-Aldrich), the protein was precipitated using either the anti-carboxy-terminal antibody or the anti-amino-terminal antibody diluted 1:1000. Medium and cell lysates were incubated for 24 hr at 4C with the respective antibody and subsequently with 20 µl of protein A-Sepharose beads. Immunoprecipitates were subjected to SDS-PAGE and immunoblot analysis.
Tissue Expression Pattern of pCLCA4a mRNA
Total RNA from organs listed above was isolated using the Trizol method (Invitrogen, Karlsruhe, Germany) and purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany), including digestion with DNase I (Qiagen, Hilden, Germany). In sum, 750 ng of RNA were reverse transcribed as described above, and the cDNA served as template in the following PCR reactions. Specific pCLCA4a primers were designed to amplify a 519-bp product (upstream 5′-GCC-GTATACCAAGCAGTTCAC-3′; 5′-CGGTGCCACCA-TTACTTCTGTGATA-3′). PCR amplification using the DreamTaq DNA Polymerase (Fermentas) included 29 cycles at 95C for 2 min, 95C for 30 sec, 57C for 30 sec, and 72C for 30 sec, followed by a final extension step at 72C for 10 min with a time increment of 1 sec per cycle. To control for mRNA quality and efficacy of reverse transcription, a 542-bp product of the mRNA coding for the housekeeping factor EF-1a was RT-PCR amplified from each sample using the primers 5′-GAACGGGCAGACCCGTGAGC-3′ (upstream) and 5′-AGCCCACGTTGTCCCCAGGA-3′ (downstream).
Primer pairs were designed to specifically discriminate among all known pCLCA homologs (Plog et al. 2009) and to encompass an intron to exclude amplification of contaminating genomic DNA. No crossreactions were detected when cloned cDNA samples of pCLCA1, pCLCA2, or pCLCA4b were used as templates.
Immunohistochemistry
Tissues were either fixed in 4% neutral buffered formaldehyde for 24 hr and embedded in paraffin or shock frozen in liquid nitrogen and stored at −80C. Paraffin-embedded tissues were cut at 3-µm thickness, mounted on adhesive glass slides, and dewaxed in xylene, followed by rehydration in descending graded ethanol. Endogenous peroxidase was blocked with 0.5% H2O2 in methanol for 30 min at room temperature. Antigen retrieval was performed using either 15 min of microwave heating (600 W) in 10 mM citric acid, pH 6.0, containing 0.05% Triton X-100 or a pretreatment with protease (AppliChem, Darmstadt) for 10 min at 37C, as well as both methods combined. Frozen tissues were cut at 5-µm thickness, mounted on adhesive glass slides, and stored at −80C until further use. Cryostate sections were fixed in acetone for 10 min, rinsed in PBS for 10 min, and peroxidase blocked as described above. The slides were washed in PBS containing 0.05% Triton X-100 and blocked with PBS containing 2% bovine serum albumin and 20% normal goat serum for 30 min. Slides were incubated overnight at 4C with anti-pCLCA4a antibodies, preimmune sera, or antibodies that had been preincubated with 50 µg/ml of the corresponding peptides or an irrelevant peptide, respectively. Antibody dilutions ranged from 1:500 to 1:10,000 with optimal results for 1:2000 for p4a-N-1b and 1:1500 for p4a-C-1a. Sections were washed repeatedly in PBS containing 0.05% Triton X-100 and incubated for 1 hr with biotinylated secondary goat anti-rabbit antibody diluted 1:200. Color was developed by incubating the slides with freshly prepared ABC solution (Vectastain Elite ABC Kit, Vector Laboratories, Inc, Burlingame, CA), followed by repeated washes and exposure to DAB (Merck, Darmstadt, Germany). Sections were counterstained with hematoxylin, dehydrated in ascending graded ethanol, cleared in xylene, and coverslipped. Consecutive sections from each tissue sample were stained with hematoxylin and eosin for histological examination. Each of these experiments was repeated at least two times.
Endoglycosidase Treatment
Cell lysates and medium were incubated with endo H or PNGase F (New England Biolabs, Frankfurt, Germany) before SDS-PAGE analysis with the antibody p4a-N-1b or p4a-C-1a, respectively. Briefly, an aliquot of each sample was incubated with 50 U/ml of endo H or 25 U/ml of PNGase F, respectively, for 1 hr at 37C. One sample was left untreated as negative control in each experiment.
Results
Genomic Structures of pCLCA4a, hCLCA4, and mClca6
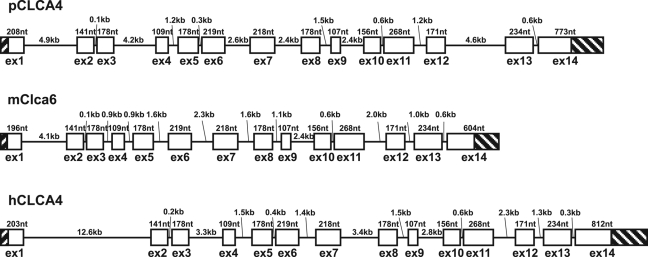
Species-specific differences in genomic structure and variability of genes are of prime importance when animal models are used for studying CF. Comparison of the exon–intron structures of pCLCA4a and its orthologs in humans and mice revealed a common structure consisting of 14 exons (Fig. 1). Regular open reading frames are present in all three orthologs, with minor differences in total protein lengths. While the pCLCA4 protein contains 927 predicted amino acids, the hCLCA4 protein is 917 amino acids long and mCLCA6, 924. Gaps in the amino acid sequence alignment were found in the terminal exons, resulting in differences in the leader peptide in exon 1 as well as the carboxy-terminus in exon 14. Both regions of the protein also show relatively low homology within the three species as well as to other CLCA4 orthologs in different mammals (data not shown). In contrast, the other regions of the protein representing 88% of the total alignment revealed high amino acid homologies to hCLCA4 and mClca6, 76.8% and 71.2%, respectively. The rate of similarity is 87.8% to hCLCA4 and 84.7% to mClca6. All exon–intron boundaries are conserved, including a rare GC-splice donor site at the 3′-end of exon 4. The functionality of this exon–intron structure is supported by the presence of a GC-donor site carboxy-terminally to exon 4 in the horse, dog, cattle, mouse, rat, man, macaque, and marmoset (data not shown).
Figure 1.
Genomic structure of pCLCA4a and its murine and human orthologs, mClca6 and hCLCA4. The three genes possess the same exon–intron structures containing 14 exons. Exon structures and the lengths of exons and introns are shown schematically, with untranslated regions shaded and coding regions displayed as open boxes. Exons are scaled 10-fold larger than introns.
Generation of Antibodies and Protein Characterization by Cellular Transport and Glycosylation Assays
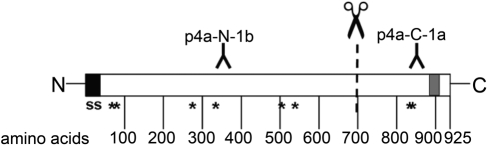
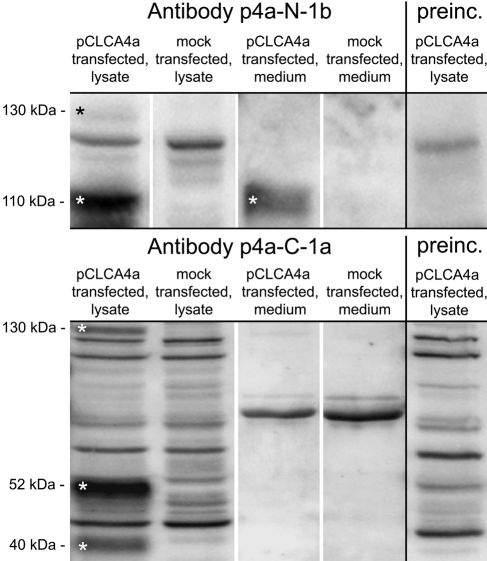
A common feature of all known CLCA proteins is their posttranslational cleavage of a precursor protein into a larger amino-terminal and a smaller carboxy-terminal product at about aa 700 (Patel et al. 2009). Thus, two different antibodies were generated (Fig. 2), one directed against an epitope within the predicted amino-terminal segment (p4a-N-1b) and one directed against an epitope within the predicted carboxy-terminal segment (p4a-C-1a). The antibody directed against the amino-terminal cleavage product (p4a-N-1b) recognized a 130-kDa precursor protein and a 110-kDa amino-terminal cleavage product on immunoblots of cell lysates from pCLCA4a-overexpressing HEK293 cells (Fig. 3, upper panel). The antibody directed against the carboxy-terminal cleavage product (p4a-C-1a) also recognized the 130-kDa precursor protein and two carboxy-terminal cleavage products of approximately 52 kDa and 40 kDa in the cell lysate (Fig. 3, bottom). Preincubation of the antibodies with their respective peptides used for immunization abolished the specific protein bands on the blots but no unspecific protein bands (Fig. 3, right panels).
Figure 2.
Protein structure of pCLCA4a, predicted transmembrane domain, and location of the peptides used for antibody generation. All software programs used predicted a transmembrane domain in the region between aa 885 and 912 (gray box) in addition to the signal sequence (ss) at the amino-terminus. NetNGlyc predicted eight potential asparagine-linked glycosylation sites (asterisks). In accordance with a predicted cleavage of the protein at about aa 700, one antibody was generated against a peptide from within the amino-terminal cleavage product (p4a-N-1b) and one against a peptide from within the carboxy-terminal cleavage product (p4a-C-1a).
Figure 3.
Only the amino-terminal subunit of pCLCA4a is secreted into the supernatant of transiently transfected HEK293 cells. The anti-amino-terminal antibody p4a-N-1b detected a pCLCA4a precursor molecule of approximately 130 kDa as well as the amino-terminal cleavage product of approximately 110 kDa in cell lysates. The amino-terminal cleavage product was also detected in the supernatant (medium). The anti-carboxy-terminal antibody p4a-C-1a detected the precursor molecule and two different carboxy-terminal cleavage products of approximately 52 and 40 kDa in the cell lysate, whereas no specific bands were detected in the supernatant. Preincubation of the antibodies with their respective peptides confirmed their specificity (right panels). Specific bands are indicated by white asterisks.
To date, CLCA proteins can be classified into one of two classes: first, fully secreted proteins, with both cleavage products being secreted by the cell; second, partially secreted proteins, with the amino-terminal protein being released by the cell and with the carboxy-terminal fragment remaining associated with the plasma membrane via an integral transmembrane domain (Patel et al. 2009). The classification of the CLCA proteins into different classes is important to draw conclusions about their possible functions as signaling molecules, components of the mucus, or transmembrane proteins possibly interacting with adjacent cells. In this study, the pCLCA4a 110-kDa amino-terminal fragment was detectable in only the supernatant by the amino-terminal antibody, whereas the carboxy-terminal cleavage products were undetectable in the supernatant of transfected HEK293 cells (Fig. 3), thereby suggesting that pCLCA4a belongs to the group of membrane-associated and only partially secreted CLCA proteins.
To further corroborate a carboxy-terminal transmembrane domain, we performed computational analyses on the predicted amino acid sequence of pCLCA4a. The SignalP 3.0 software identified a single hydrophobic, amino-terminal, cleavable signal sequence, most likely between aa 20 and 21 (Fig. 2). In addition, all six programs used for transmembrane prediction consistently predicted one carboxy-terminal transmembrane domain between aa 885 and 912 (Fig. 2, gray box). The DAS program revealed an additional transmembrane domain between aa 617 and 627. For prediction of N-linked glycosylation sites within the pCLCA4a protein, we used the NetNGly software, which identified eight consensus glycosylation sites at aa 74, 85, 282, 337, 501, 533, 829, and 834, with six located in the larger amino-terminal part of the protein and two in the smaller carboxy-terminal part (Fig. 2).
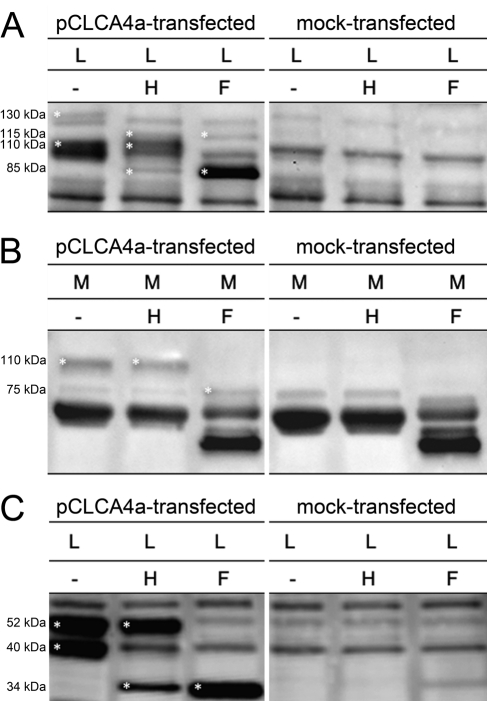
Cellular transport assays of the murine ortholog to pCLCA4a, mCLCA6, have previously identified a cleavage of the CLCA precursor protein in the endoplasmic reticulum, followed by a transport of the cleaved products through the Golgi apparatus where they become complex glycosylated (Bothe et al. 2008). To test whether pCLCA4a is processed and transported similarly, the cell lysate and medium of pCLCA4a-transfected HEK293 cells were treated with the endoglycosidases endo H and PNGase F, respectively, and immunoblotted using either the anti-amino-terminal or the anti-carboxy-terminal antibody. In the cell lysate, the 130-kDa precursor protein was sensitive to endo H and PNGase F, resulting in a reduction in size by approximately 15 kDa (Fig. 4A, left panel). Obviously, the precursor protein has not passed the Golgi and is cleaved in the endoplasmic reticulum as an immature glycoprotein with high-mannose-type glycosylation. For the amino-terminal cleavage product, a small proportion was found to be endo H sensitive, but the major part was sensitive to only PNGase F. The amino-terminal cleavage product detected in the medium was sensitive to PNGase F only, consistent with a secreted protein of complex glycosylation (Fig. 4B, left).
Figure 4.
Posttranslational processing of pCLCA4a indicates proteolytic cleavage in the endoplasmic reticulum and complex glycosylation of both subunits. (A) The precursor molecule in the cell lysate (L) was sensitive to both endo H (H) and PNGase F (F) as indicated by a shift in size from approximately 130 kDa to approximately 115 kDa. In contrast, the amino-terminal cleavage product was only partially sensitive to endo H but was mainly deglycosylated by PNGase F as shown by a shift of approximately 25 kDa. (B) In the supernatant (medium, M), the amino-terminal cleavage product was only deglycosylated by PNGase F but not by endo H, indicating a complex glycosylation pattern. (C) In the cell lysate, the approximately 52-kDa carboxy-terminal subunit was sensitive to PNGase F only, as shown by a shift to approximately 34 kDa, whereas the approximately 40-kDa carboxy-terminal subunit was also deglycosylated by endoH, suggesting an immature glycosylation of the latter. Specific bands are indicated by asterisks.
The anti-carboxy-terminal antibody (p4a-C-1a) detected a 130-kDa precursor protein sensitive to endo H and PNGase F and thus confirmed the data obtained with the antibody p4a-N-1b (not shown). The 40-kDa carboxy- terminal cleavage product was reduced to a 34-kDa fragment by endo H and PNGase F, indicating that this cleavage product was mannose-rich glycosylated, located in the endoplasmic reticulum. In contrast, the 52-kDa carboxy-terminal cleavage product was sensitive to PNGase F treatment only and shifted to a 34-kDa protein (Fig. 4C, left panel). Obviously, this cleavage product is a complex glycosylated form that has reached the Golgi apparatus. Thus, the cellular transport of pCLCA4a appears similar to that of its murine ortholog mCLCA6 (Bothe et al. 2008). Specificity of protein bands was confirmed by including extracts from mock-transfected cells and medium in each experiment (Fig 4A–C, right panels).
Tissue and Cellular Distribution Patterns of pCLCA4a
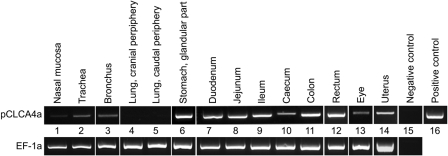
The expression patterns of orthologous CLCA family members may differ between species and should be transfered with caution from one species to another. Bothe et al. (2008) have shown that the murine mCLCA6 is exclusively expressed in the intestinal tract, which stands in sharp contrast to the widespread expression pattern of its human ortholog hCLCA4, including the lung. To establish the expression pattern of their ortholog in the pig, pCLCA4a, we performed systematic expression analyses on the mRNA and protein levels. pCLCA4a mRNA was found in the entire intestinal tract, in the glandular part of the stomach, in all parts of the respiratory tract that contain respiratory-ciliated epithelium, in the uterus, and in the eye (Fig. 5).
Figure 5.
Broad expression pattern of pCLCA4a on the mRNA level. Using pCLCA4a-specific primers, mRNA was detected not only in the gastrointestinal tract (lanes 6 to 12) but also in the upper respiratory tract (lanes 1 to 3), the eye (lane 13), and the uterus (lane 14). Of note, pCLCA4a mRNA was not detected in samples from the lung parenchyma without ciliated epithelium (lanes 4 and 5). A single specific band of 519 base pairs was detected when the pCLCA4a clone was used as template (lane 16). A 542-bp fragment of the mRNA coding for the housekeeping gene EF-1a was RT-PCR amplified to control for RNA integrity and efficacy of reverse transcription (bottom line). The figure shown is representative for two independent experiments in three different animals.
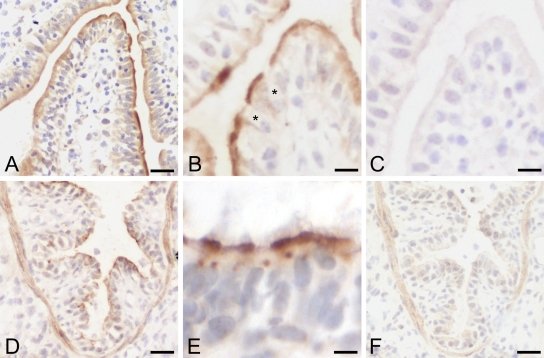
Immunohistochemical analyses using the two antibodies, p4a-N-1b and p4a-C-1a, yielded virtually identical results for all tissues and animals tested. Despite different antigen retrieval methods and different antibody dilutions, the pCLCA4a protein could not be detected on formalin-fixed, paraffin-embedded tissues. Hence, all experiments were conducted on cryostate sections (Fig. 6). Both antibodies generated against either the predicted amino-terminal cleavage product (p4a-N-1b) or the predicted carboxy- terminal cleavage product (p4a-C-1a) resulted in identical staining patterns, indicating strict colocalization of both cleavage products. Strong and consistent staining for the pCLCA4a protein was observed with both antibodies in the apical membranes of non–goblet cell enterocytes of the enteric villus tips throughout the small intestine (Fig. 6A, B). Specific staining was not observed in the deep crypt cells or in any of the large intestinal segments. The pCLCA4a protein was also detected in the respiratory tract from the nasal cavity down to the small bronchi with apical membranes of virtually all respiratory epithelial cells strongly stained (Fig. 6D). Occasionally, the protein was also detected adjacent to the nucleus, consistent with its suggested presence in the endoplasmic reticulum during posttranslational modification (Fig. 6E). Staining of bronchial smooth muscle cells was considered unspecific because virtually identical staining was observed with the preimmune sera (Fig. 6F). This assumption was supported by a similar staining of smooth muscle cells in other organs independent of any pCLCA4a mRNA detection—for example, in the small intestine or the urinary bladder (not shown). In contrast to that in the intestinal tract, the pCLCA4a protein was detected in non-goblet and goblet cells of the respiratory epithelium. No protein was detected in the alveolar walls, the interstitium, serosal linings, or lymphatic tissues of the lungs. The submucosal glands were either unstained by immunohistochemistry or showed unspecific background that was also present in sections incubated with preimmune sera or after precincubation of the specific antibodies with the antigenic peptide (not shown). Although pCLCA4a mRNA was detected in the stomach, in the uterus, and in the eye, no immunohistochemical signals were observed in these organs (not shown).
Figure 6.
The pCLCA4a protein is located apically in intestinal and respiratory epithelial cells. In the small intestine, the pCLCA4a protein was primarily detected along the apical membranes of non–goblet cell enterocytes on the villus tips (A, B). Goblet cells were spared from staining (black asterisks, B). In the respiratory tract, pCLCA4a was localized in epithelial cells (D) with a strong apical membrane staining pattern and occasional spotlike signals adjacent to the nucleus (E). Signals within smooth muscle cells of the bronchi were considered unspecific due to an identical staining pattern given by the preimmune sera (F). Sections stained with preimmune sera failed to reveal any specific signals (C, F). Immunohistochemistry using antibody p4a-N-1b diluted 1:4000. Bars: (A, D, F) 25 µm, (B, C) 30 µm, (E) 15 µm.
Staining with preimmune sera or immune sera from rabbits immunized with non-pCLCA4a-related antigens confirmed specificity of the pCLCA4a signals (Fig. 6C, F). After preincubation of the antibodies with the peptides used for immunization, either the signals disappeared, or their intensities were strongly reduced, whereas preincubation of the antibodies with an irrelevant peptide failed to reduce signal intensities (not shown).
Immunoblotting and Immunoprecipitation
Despite clear and consistent specific signals with both antibodies on immunoblots of lysates from pCLCA4a-overexpressing HEK293 cells, none of the antibodies detected a specific band on immunoblots of whole tissue lysates. In a second step, each blot membrane was stripped and reprobed with an anti-β-actin antibody, confirming protein integrity. Even after immunoprecipitation of cell lysates from organs that had a specific signal in immunohistochemistry, no specific bands were detected by either of the antibodies used, despite clear and consistent bands after immunoprecipitation of pCLCA4a-overexpressing HEK293 cells (not shown). It was thus concluded that the overall protein expression levels were too low for their detection on immunoblots from whole organ lysates.
Discussion
The generation of a porcine CF model that displays the human CF pathology much more closely than murine models do (Rogers, Stoltz, et al. 2008; Meyerholz, Stoltz, Namati et al. 2010; Meyerholz, Stoltz, Pezzulo et al. 2010; Stoltz et al. 2010) opens new possibilities to understand the mechanisms of this fatal disease and to develop and test new therapeutic strategies. In addition to the CFTR genotype, environmental factors as well as modifier genes are important determinants of the CF phenotype (Collaco and Cutting 2008). Among others, members of the CLCA gene family, especially hCLCA1 and hCLCA4 as well as their murine orthologs mCLCA3 and mCLCA6, respectively, are thought to contribute to the distinct phenotypes of CF in humans and mouse models, respectively (Ritzka et al. 2004; Young et al. 2007; Bothe et al. 2008; van der Doef et al. 2010). In view of the new porcine CF model, we aimed at characterizing pCLCA4a—the porcine ortholog to the human hCLCA4 and the murine mCLCA6—in terms of its gene structure, tissue expression pattern, and cellular protein processing, focusing on possible differences to and similarities from its human and murine orthologs.
The tissue expression patterns of pCLCA4a and its human and murine orthologs are illustrated in Table 1, indicating that pCLCA4a has more similarities to the human hCLCA4 than to the murine mCLCA6. The major difference between pCLCA4a and mCLCA6, which has been detected exclusively in the intestine (Bothe et al. 2008), is the detection of pCLCA4a mRNA and protein in respiratory epithelia. Importantly, this is fully consistent with the human ortholog hCLCA4 (Agnel et al.1999; Mall et al. 2003). The pCLCA4a protein was found in virtually all respiratory epithelial cells from the nasal cavity down to the small bronchi but without any expression in the distal lung parenchyma, including type I or II pneumocytes, or in the submucosal glands. In addition, the pCLCA4a protein was found in the apical membranes of the small intestinal villus epithelial cells, similar to its murine ortholog mCLCA6 (Bothe et al. 2008). Expression of the pCLCA4a protein on the apical membranes of the respiratory and intestinal tracts would be consistent with a role in transepithelial ion conduction as proposed for several CLCA members (Evans et al. 2004; Hamann et al. 2009). Further studies will have to address the functional role of this protein at the apical membranes of respiratory and intestinal epithelia.
Table 1.
Comparison of the Expression Patterns of the Porcine, Human, and Murine Orthologs pCLCA4a, hCLCA4, and mCLCA6
| pCLCA4a |
hCLCA4 |
mCLCA6 |
||||
|---|---|---|---|---|---|---|
| Tissue | RNA | Protein | RNA | Protein | RNA | Protein |
| Nasal mucosa | + | + | +b | ND | −d | −d |
| Trachea | + | + | +a | ND | −d | −d |
| Bronchi | + | + | ND | ND | −d | −d |
| Lung (nonbronchial) | − | − | −a | ND | −d | −d |
| Heart | − | − | −a | ND | −d | −d |
| Aorta | − | − | −a | ND | ND | ND |
| Oesophagus | − | − | ND | ND | −d | −d |
| Stomach, nonglandular part | − | − | ND | ND | −d | −d |
| Stomach,glandular part | + | − | +a | ND | − / + d/c | −d |
| Small intestine | + | + | +a | ND | +c,d | +d |
| Large intestine | + | − | +a | ND | +c,d | +d |
| Gall bladder | − | − | ND | ND | −d | −d |
| Liver | − | − | −a | ND | − / + c,d | −d |
| Spleen | − | − | −a | ND | − / + c,d | −d |
| Kidney | − | − | −a | ND | −d | − d |
| Urinary bladder | − | − | +a | ND | −d | −d |
| Pancreas | − | − | −a | ND | −d | −d |
| Salivary gland | − | − | +a | ND | −d | − d |
| Eye | + | − | ND | ND | − / + c,d | −d |
| Mandibular lymph node | − | − | −a | ND | −d | −d |
| Brain | − | − | +a | ND | −d | −d |
| Pituitary gland | − | − | −a | ND | ND | ND |
| Thoracic ganglion | − | − | ND | ND | ND | ND |
| Skin | − | − | ND | ND | −d | −d |
| Testicle | − | − | +a | ND | −d | −d |
| Epididymis | − | − | ND | ND | −d | −d |
| Vas deferens / spermatic cord | − | − | ND | ND | ND | ND |
| Prostate gland | − | − | +a | ND | ND | ND |
| Uterus | + | − | +a | ND | −d | −d |
| Ovaries | − | − | −a | ND | −d | −d |
+, detection of RNA/protein; −, no detection of RNA/protein; ND, not done.
Of additional interest is the comparison of the pCLCA4a protein expression with that of the porcine CFTR protein. In fact, the respiratory cellular expression pattern of pCLCA4a is virtually identical to that of the porcine CFTR protein (Plog et al. 2010). A modulatory role of pCLCA4a in the pathogenesis of the CF respiratory disease or other diseases of the respiratory tract is therefore conceivable. Furthermore, a recently published study of CF pig models that carry the common CFTR-Δ508 mutation revealed a partial transport of the porcine CFTR protein to the respiratory apical membrane, resulting in a residual CFTR conductance of 6% compared with the wild-type animals (Ostedgaard et al. 2011). Despite this residual conductance, these pigs develop the characteristic intestinal and lung disease. It would be of prime interest to compare the localization and quantity of pCLCA4a in the CFTRΔ508/Δ508 pigs with a wild-type control group. Future studies are needed to examine differences in the expression pattern of pCLCA4a in these promising new animal models of CF and to investigate a possible functional or regulating interaction between the mutated pCFTR protein and pCLCA4a.
In contrast to the respiratory tract, the cellular expression patterns of pCLCA4a and pCFTR in the intestine appear to be slightly different. Both proteins are expressed in enterocytes, but the pCFTR protein has been found predominantly in the crypts (Plog et al. 2010), which fail to express the pCLCA4a protein. The consequences of this partially overlapping expression patterns will have to be addressed in future studies on the pig model of CF.
Interestingly, pCLCA4a exhibits a broader tissue expression pattern on the mRNA level than on the protein level. In contrast to strong mRNA expression in the stomach, large intestine, eye, and uterus, no pCLCA4a protein was detected in these tissues. A similar discrepancy has been reported for other CLCA members. For example, the murine mCLCA5 mRNA was found in the corneal epithelium without detectable protein expression (Braun et al. 2010). As a possible explanation, a posttranscriptional regulatory event preventing protein synthesis in specific tissues might be responsible. Alternatively, a lower sensitivity of the immunohistochemical assay must be taken into account, compared to the highly sensitive RT-PCR. It is important to note that the pCLCA4a protein was only detectable on cryostate sections but not on formalin-fixed, paraffin-embedded tissues in light of the fact that immunohistochemistry on cryostate sections is generally known to be more sensitive. The pCLCA4a protein may therefore be undetectable in tissues with low protein abundance using immunohistochemistry. The lack of pCLCA4a detection on immunoblots of whole tissue lysates supports this notion.
In terms of its genomic structure, protein structure, and protein processing, the porcine pCLCA4a is very similar to its human und murine orthologs. The genomic structure of the pCLCA4a gene did not significantly differ from its human und murine orthologs. The number of exons and their distribution indicate a close synteny among the three species. Similar to its murine ortholog mCLCA6, the pCLCA4a protein can be assigned to the CLCA group of heterodimeric glycoproteins of which only the larger amino-terminal subunit is secreted by the cell, whereas the smaller carboxy-terminal subunit is anchored to the plasma membrane by an integral transmembrane domain. In this regard, pCLCA4a is similar to its murine ortholog mCLCA6. Also similar to mCLCA6 (Bothe et al. 2008), the protein is cleaved within the endoplasmic reticulum, and the cleaved protein products reach the Golgi apparatus, where they become complex glycosylated. Unfortunately, experimental data on the protein processing of the human hCLCA4 are not available to date. However, in silico analyses of its amino acid sequence argue that hCLCA4 can also be assigned to the group of CLCA proteins with a transmembrane domain (data not shown). Thus, with respect to the cellular protein processing and structure, there seem to be no considerable differences between pCLCA4a and its human and murine orthologs.
In summary, the genomic organization, overall protein structure, and protein processing of pCLCA4a appear to be similar to its human and murine orthologs hCLCA4 and mCLCA6, respectively. However, considerable species-specific differences exist in their tissue and cellular expression patterns, with closer homology of the lung expression of pCLCA4a to the human hCLCA4 than to the murine mCLCA6. This argues that the novel porcine CF model may be more suitable than CF mouse models for studying the proposed modulatory role of hCLCA4.
Acknowledgments
This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft MU 3015/1-1). This work is part of the doctoral theses of SP and TG.
Footnotes
The authors declared no potential conflicts of interest with respect to the authorship and publication of this article.
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft MU 3015/1-1).
References
- Agnel M, Vermat T, Culouscou JM. 1999. Identification of three novel members of the calcium-dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett. 455:295–301 [DOI] [PubMed] [Google Scholar]
- Anton F, Leverkoehne I, Mundhenk L, Thoreson WB, Gruber AD. 2005. Overexpression of eCLCA1 in small airways of horses with recurrent airway obstruction. J Histochem Cytochem. 53:1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe MK, Braun J, Mundhenk L, Gruber AD. 2008. Murine mCLCA6 is an integral apical membrane protein of non- goblet cell enterocytes and co-localizes with the cystic fibrosis transmembrane conductance regulator. J Histochem Cytochem. 56:495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J, Bothe MK, Mundhenk L, Beck CL, Gruber AD. 2010. Murine mCLCA5 is expressed in granular layer keratinocytes of stratified epithelia. Histochem Cell Biol. 133:285–299 [DOI] [PubMed] [Google Scholar]
- Collaco JM, Cutting GR. 2008. Update on gene modifiers in cystic fibrosis. Curr Opin Pulm Med. 14:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon CJ, Yamasaki K, Kawasaki S, Quantock AJ, Koizumi N, Kinoshita S. 2004. Calcium-activated chloride channel-2 in human epithelia. J Histochem Cytochem. 52:415–418 [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Awayda MS, Bubien JK, Ismailov, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. 1995. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. 270:31016–31026 [DOI] [PubMed] [Google Scholar]
- Davidson DJ, Rolfe M. 2001. Mouse models of cystic fibrosis. Trends Genet. 17:S29–S37 [DOI] [PubMed] [Google Scholar]
- Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU. 2006. The putative chloride channel hCLCA2 has a single c-terminal transmembrane segment. J Biol Chem. 281:29448–29454 [DOI] [PubMed] [Google Scholar]
- Evans SR, Thoreson WB, Beck CL. 2004. Molecular and functional analyses of two new calcium-activated chloride channel family members from mouse eye and intestine. J Biol Chem. 279:41792–41800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar KJ, Racette KJ, Gordon JR, Loewen ME, Forsyth GW. 2000. Cloning a chloride conductance mediator from the apical membrane of porcine ileal enterocytes. Physiol Genomics. 3:101–111 [DOI] [PubMed] [Google Scholar]
- Gandhi R, Elble RC, Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU. 1998. Molecular and functional characterization of a calcium-sensitive chloride channel from mouse lung. J Biol Chem. 273:32096–32101 [DOI] [PubMed] [Google Scholar]
- Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, Thompson N. 2005. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J Biol Chem. 280:27205–27212 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Elble RC, Ji HL, Schreur KD, Fuller CM, Pauli BU. 1998. Genomic cloning, molecular characterization, and functional analysis of human CLCA1, the first human member of the family of Ca2+-activated Cl- channel proteins. Genomics. 54:200–214 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Gandhi R, Pauli BU. 1998. The murine calcium-sensitive chloride channel (mCaCC) is widely expressed in secretory epithelia and in other select tissues. Histochem Cell Biol. 110: 43–49 [DOI] [PubMed] [Google Scholar]
- Hamann M, Gibson A, Davies N, Jowett A, Walhin JP, Partington L, Affleck K, Trezise D, Main M. 2009. Human ClCa1 modulates anionic conduction of calcium dependent chloride currents. J Physiol. 587(Pt 10):2255–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 14:378–379 [DOI] [PubMed] [Google Scholar]
- Jeong SM, Park HK, Yoon IS, Lee JH, Kim JH, Jang CG, Lee CJ, Nah SY. 2005. Cloning and expression of Ca2+-activated chloride channel from rat brain. Biochem Biophys Res Commun. 334:569–576 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Leverkoehne I, Gruber AD. 2002. The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory, and uterine goblet cells. J Histochem Cytochem. 50:829–838 [DOI] [PubMed] [Google Scholar]
- Loewen ME, Forsyth GW. 2005. Structure and function of CLCA proteins. Physiol Rev. 85:1061–1092 [DOI] [PubMed] [Google Scholar]
- Loewen ME, Gabriel SE, Forsyth GW. 2002. The calcium-dependent chloride conductance mediator pCLCA1. Am J Physiol Cell Physiol. 283:C412–C421 [DOI] [PubMed] [Google Scholar]
- Mall M, Gonska T, Thomas J, Schreiber R, Seydewitz HH, Kuehr J, Brandis M, Kunzelmann K. 2003. Modulation of Ca2+-activated Cl- secretion by basolateral K+ channels in human normal and cystic fibrosis airway epithelia. Pediatr Res. 53:608–618 [DOI] [PubMed] [Google Scholar]
- Meyerholz DK, Stoltz DA, Namati E, Ramachandran S, Pezzulo AA, Smith AR, Rector MV, Suter MJ, Kao S, McLennan G, et al. 2010. Loss of cystic fibrosis transmembrane conductance regulator function produces abnormalities in tracheal development in neonatal pigs and young children. Am J Respir Crit Care Med. 182:1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz DK, Stoltz DA, Pezzulo AA, Welsh MJ. 2010. Pathology of gastrointestinal organs in a porcine model of cystic fibrosis. Am J Pathol. 176:1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundhenk L, Alfalah M, Elble RC, Pauli BU, Naim HY, Gruber AD. 2006. Both cleavage products of the mCLCA3 protein are secreted soluble proteins. J Biol Chem. 281:30072–30080 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 24:34–36 [DOI] [PubMed] [Google Scholar]
- Nielson H, Engelbrecht J., Brunack S., von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- Ostedgaard LS, Meyerholz DK, Chen JH, Pezzulo AA, Karp PH, Rokhlina T, Ernst SE, Hanfland RA, Reznikov LR, Ludwig PS, et al. 2011. The ΔF508 mutation causes CFTR misprocessing and cystic fibrosis-like disease in pigs. Sci Transl Med. 3:74ra24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Rogers CS, Dong Q, Randak CO, Vermeer DW, Rokhlina T, Karp PH, Welsh MJ. 2007. Processing and function of CFTR-DeltaF508 are species-dependent. Proc Natl Acad Sci U S A. 104:15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AC, Brett TJ, Holtzman MJ. 2009. The role of CLCA proteins in inflammatory airway disease. Annu Rev Physiol. 71:425–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CT, Huang W, Jarrahy R, Rudkin G, Yamaguchi DT, Miller TA. 2009. Human and mouse osteoprogenitor cells exhibit distinct patterns of osteogenesis in three-dimensional tissue engineering scaffolds. Plast Reconstr Surg. 124:1869–1879 [DOI] [PubMed] [Google Scholar]
- Plog S, Mundhenk L, Bothe MK, Klymiuk N, Gruber AD. 2010. Tissue and cellular expression of porcine CFTR: similarities to and differences from human CFTR. J Histochem Cytochem. 58:785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plog S, Mundhenk L, Klymiuk N, Gruber AD. 2009. Genomic, tissue expression, and protein characterization of pCLCA1, a putative modulator of cystic fibrosis in the pig. J Histochem Cytochem. 57:1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzka M, Weinel C, Stanke F, Tümmler B. 2004. Sequence comparison of the whole murine and human CLCA locus reveals conserved synteny between both species. Genome Lett. 2:149–154 [Google Scholar]
- Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB, Jr, McLennan G, Meyerholz DK, Namati E, Ostedgaard LS, et al. 2008. The porcine lung as a potential model for cystic fibrosis. Am J Physiol Lung Cell Mol Physiol. 295:L240–L263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, Rogan MP, Pezzulo AA, Karp PH, Itani OA, et al. 2008. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 321:1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte BJ, Davidson DJ, Wilke M, De Jonge HR. 2004. Animal models of cystic fibrosis. J Cyst Fibros. 3 Suppl 2:183–190 [DOI] [PubMed] [Google Scholar]
- Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA, et al. 2010. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci Transl Med. 2:29ra31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics. 17:849–850 [DOI] [PubMed] [Google Scholar]
- van der Doef HP, Slieker MG, Staab D, Alizadeh BZ, Seia M, Colombo C, van der Ent CK, Nickel R, Witt H, Houwen RH. 2010. Association of the CLCA1 p.S357N variant with meconium ileus in European patients with cystic fibrosis. J Pediatr Gastroenterol Nutr. 50:347–349 [DOI] [PubMed] [Google Scholar]
- Young FD, Newbigging S, Choi C, Keet M, Kent G, Rozmahel RF. 2007. Amelioration of cystic fibrosis intestinal mucous disease in mice by restoration of mCLCA3. Gastroenterology. 133:1928–1937 [DOI] [PubMed] [Google Scholar]
- Zitnik RJ, Zhang J, Kashem MA, Kohno T, Lyons DE, Wright CD, Rosen E, Goldberg I, Hayday AC. 1997. The cloning and characterization of a murine secretory leukocyte protease inhibitor cDNA. Biochem Biophys Res Commun. 232:687–697 [DOI] [PubMed] [Google Scholar]