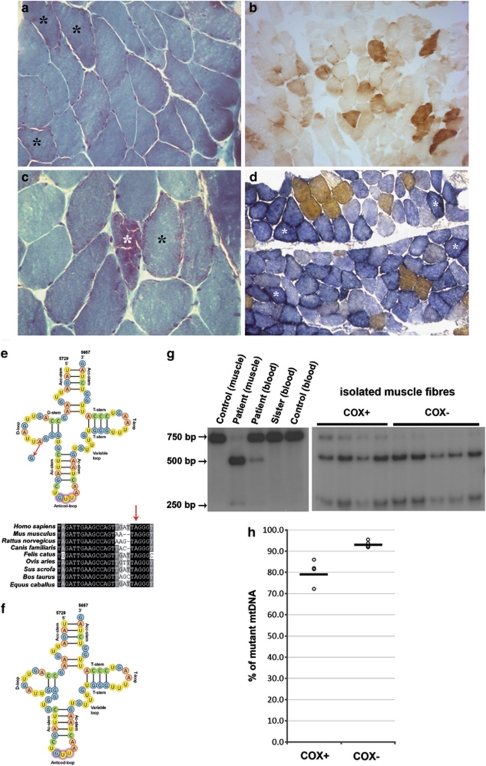
Figure 1.
Histological and histochemical findings in proband's muscle (a–d). (a) Original magnification × 200) and (c) × 400; Gomori trichrome histology showing RRFs (*) within a conserved muscle structure; (b, d × 100); COX (b) and combined COX-SDH (d) histochemistry showing severe and diffuse COX-deficiency with evidence of mitochondrial proliferation (*). (e) Cloverleaf structure of the human mitochondrial tRNAAsn (top); the m.5709T>C mutation resulting in A-to-G substitution at the RNA level is indicated by the arrow. Homologous sequences of part of the mitochondrial tRNAAsn in different species show a wide conservation of the affected nucleotide (bottom). All bases shown in black are 100% conserved in mammalian tRNAAsn genes. (f) A speculation of the mutated molecule secondary structure shows how it would likely result in a D-stem and -loop defective tRNA. (g) PCR–RFLP analysis of the m.5709T>C mutation. Autoradiogram showing BstXI-digested PCR products electrophoresed through a 5% non-denaturing polyacrylamide gel, performed on isolated fibres and on tissues from proband and controls. (h) Distribution of the mutation load measured in COX-positive (COX+, n=4) and COX-negative (COX−, n=5) muscle fibres after PCR–RFLP and densitometric analysis. Horizontal bars represent the calculated means.