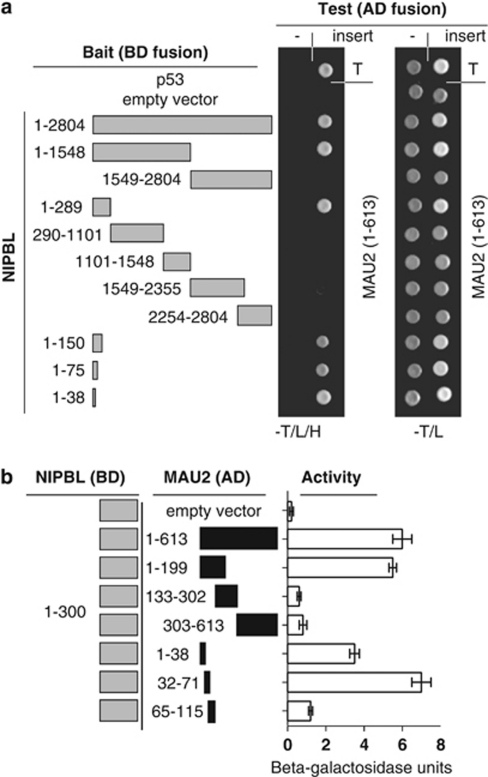
Figure 1.
Localization of NIPBL–MAU2 interaction. (a) NIPBL fragments used in confirmation and localization of NIPBL binding to MAU2 are demonstrated in gray to the left. Amino-acid residues included are indicated. The right panel demonstrates yeast two-hybrid colony assays, indicating interaction-dependent growth on tryptophan-, leucine- and histidine-deficient media (−T/L/H) of the positive control p53 with SV40TAg (T) in the uppermost row. Interaction-independent growth on tryptophan- and leucine-deficient plates is indicated to the right. NIBPL clones were tested with empty (-) and MAU2 containing AD fusion vectors. (b) The left panel depicts the NIPBL (aa 1–300) and MAU2 deletion constructs used for liquid β-galactosidase assay. The right panel indicates the interaction of the NIPBL/delangin fragment (1–300) and the different MAU2 protein fragments by liquid β-galactosidase assay.