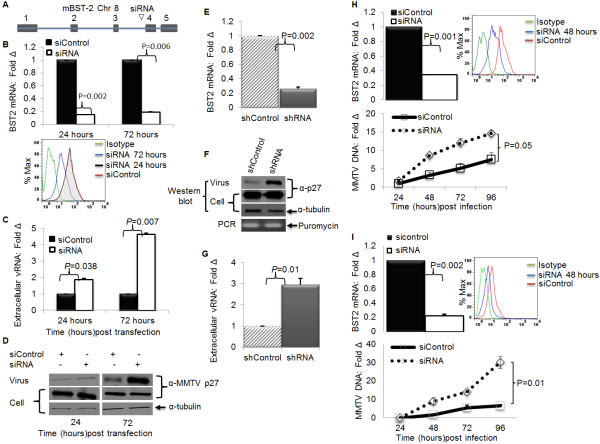
Figure 4.
Depletion of endogenous BST-2 enhances MMTV particle release and virus replication in tissue culture cells. GR cells were transfected with siRNA targeting the mouse BST-2 gene or non-targeting control siRNA (siControl). At 24 or 72 hours after transfection, both culture supernatants and their corresponding cells were collected. Virions were purified from supernatants, and both virions and corresponding cells were used for RNA and protein extractions. Aliquots of the supernatants were used to quantify the amount of extracellular virus. (A) Location of the siRNA sequence on the BST-2 gene; (B) Level of BST-2 following siRNA treatment (upper panel, bars- BST-2 mRNA presented as fold change relative to the control and lower panel, histogram- BST-2 protein); (C) Level of extracellular viral RNA following treatment with BST-2 siRNA and siControl, presented as fold change relative to the control; (D) Western blot of extracellular and intracellular viral proteins detected by antibody to the capsid protein p27; tubulin is loading control. Next, GR cells were stably transduced with shRNA targeting mouse BST-2 mRNA or a non-targeting shRNA. (E) Cells were used to determine extent of BST-2 knockdown by qPCR. (F and G) Culture supernatants and their corresponding cells were used to examine level of (F) extracellular and intracellular viral proteins by Western blot, stable integration of lentiviral particles carrying shRNA was evaluated by puromycin PCR; (G) extracellular viral RNA. To evaluate the effect of BST-2 on MMTV replication, NMuMG cells or 3T3 MEF cells were transfected with siRNA or siControl. Twenty four hours after transfection, cells were infected with MMTV and harvested at different times (shown on the figures) for DNA extraction and qPCR analysis of viral DNA, (H) qPCR analysis of endogenous NMuMG BST-2 mRNA (top-left panel), cell surface expression (top-right panel), MMTV DNA (bottom-left panel); (I) qPCR analysis of endogenous 3T3 MEF BST-2 mRNA (top-left panel), cell surface expression (top-right panel), MMTV DNA (bottom-left panel) following knockdown with siRNA or siControl. Data are normalized to GAPDH and presented as fold change relative to siControl treated cells. Error bars are standard deviation, p is significance level. Experiments were repeated at least three different times with similar results.