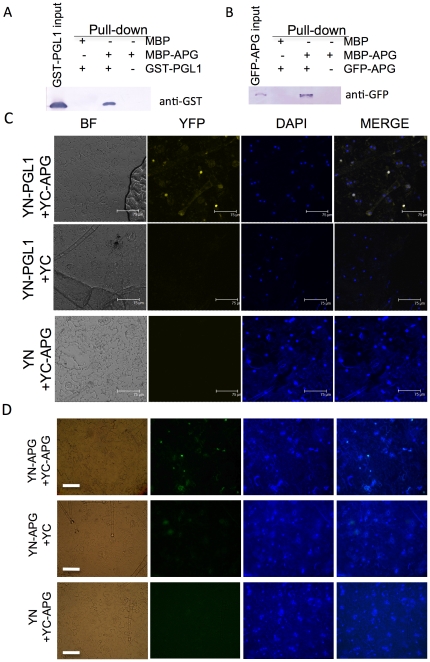
Figure 2. Interaction between PGL1 and APG.
A) Interaction between PGL1 and APG in vitro detected by pull-down assay. Amylose resin–bound MBP-APG or MBP was incubated with an equal amount of GST-PGL1. Proteins co-precipitated with the amylose resin were detected by immunoblotting using anti-GST antibody. B) In vitro homodimerization of APG detected by pull-down assay. Amylose resin–bound MBP-APG or MBP was incubated with an equal amount of GFP-APG extracted from N. benthiamiana leaves. Proteins co-precipitated with the amylose resin were detected by immunoblotting using anti-GFP antibody. C) Confocal images of interaction in vivo between PGL1 and APG revealed by BiFC assay in N. benthiamiana leaf epidermis. BF, bright field image; YFP, yellow fluorescent protein; DAPI, 4′,6-diamidino-2 phenylindole for nuclear staining; MERGE, merged view of the YFP and DAPI images. YN-PGL1+YC-APG indicates Agrobacterium mediated co-infiltration of constructs encoding N-EYFP-PGL1 and C-EYFP-APG (upper); YN-PGL1+YC, co-infitration of N-EYFP-PGL1 and C-EYFP alone (middle); YN+YC-APG, co-infiltration of N-EYFP alone and C-EYFP-APG (lower). (bar = 75 µm) D) Light microscopic images of homodimerization in vivo of APG revealed by BiFC assay. YFP was detected in the nucleus when YN-APG was co-infiltrated with YC-APG, suggesting that the protein forms a homodimer to reconstruct the YFP signal (upper). In contrast, the YFP signal was not detected when YN-APG or YC-APG was used alone. (bar = 50 µm).