Figure 2.
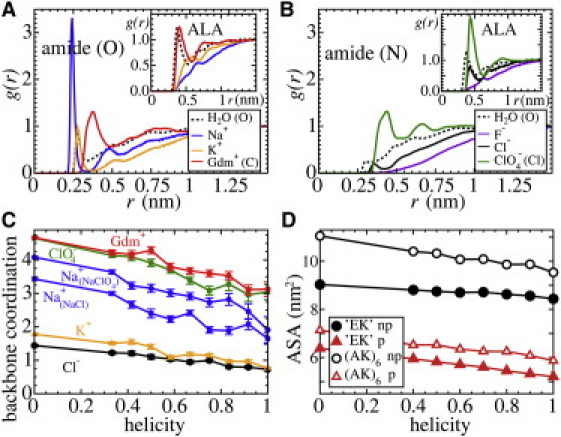
Salt-specific structures studied by MD simulations for T = 300 K. (A) Normalized density distribution g(r) of water (O) and cations (all from chloride salts) around the amide oxygen of the EK-peptide. Inset: g(r) for water and cations around the ALA methyl group. (B) Normalized density distribution g(r) of water (O) and anions around the amide nitrogen of the EK-peptide. Inset left: g(r) for water and anions around the ALA methyl group. (C) Ion coordination in a 0.5 nm shell around the backbone plotted versus average helicity h. Coordination increases upon unfolding due to direct binding to the backbone. (D) Nonpolar and polar ASA of the EK and (AK)6 peptides plotted versus helicity h. Both nonpolar and polar ASA linearly increase upon unfolding.
