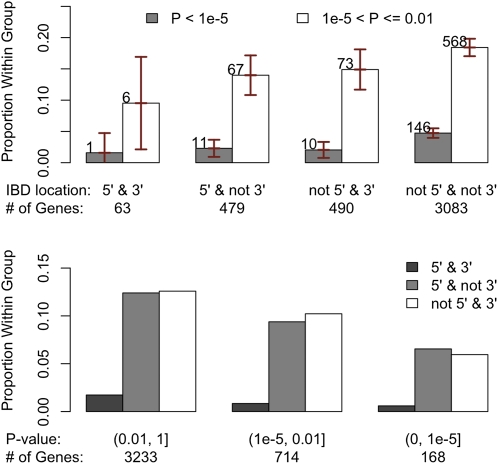
Figure 6 .
Comparisons of the categories of transcripts based on strain effect P-value at forebrain and IBD status at 5′ and 3′ regions for the 4118 transcripts with variable genomic sequence similarity estimates between NOD/ShiLtJ and C57BL/6J in the gene body. Here IBD is defined as genotypic identity > 0.999 using Sanger sequencing data. In the upper panel, the transcripts were grouped on the basis of IBD location and within each group we compared proportion of transcripts within different P-value ranges. The vertical bars indicated the 95% confidence intervals. In the lower panel, the transcripts were grouped on the basis of strain effect P-values and within each group, we compared the proportion of transcripts with different IBD location statuses.