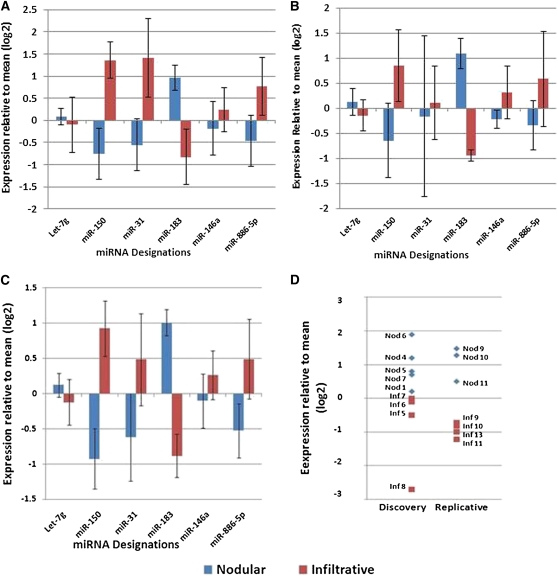
Figure 4 .
Validation of differentially expressed miRNAs by qPCR. Six miRNAs were assessed by Taqman assays. Assays for miR-141 and miR-582-5p failed because of low expression levels (see Materials and Methods). A) Among nine tumors from the discovery set in which adequate RNA was available, qPCR mirrored RNAseq in the direction and approximate magnitude of differences between the two tumor types except for Let-7g. (B) A replication set of seven tumors showed concordance in direction with qPCR results in the discovery set. (C) Upon combining the analytical and biological validation sets, three of six miRNAs, miR-150, 183, and 886-5p, were significantly different (P < 0.05) and two others, miR-31 and 146, trended in the same direction as shown by RNA sequencing but failed to reach significance. (D) Expression of miR-183 showed no overlap in nodular tumors compared with infiltrative tumors in both the discovery and replication set. Y-axis values are provided in log(2) such that a value of one indicates a twofold difference between samples.