Abstract
Hemorrhagic fever with renal syndrome (HFRS) is a serious public health issue in Far East Russia. Two different hantaviruses were isolated from rodents captured in the Khabarovsk region: Amur virus (AMRV; Khekhtsir/AP209/2005 strain from Apodemus peninsulae) and Hantaan virus (HTNV; Galkino/AA57/2002 strain from A. agrarius). Genetic analysis of the new isolates revealed that the M and L segments were apparently different between AMRV and HTNV, but S segments of the two viruses were closer. The antigenicities of AMRV, HTNV, and Seoul virus (SEOV) were differentiated by cross-neutralization. Serological differential diagnoses of 67 HFRS patients in the Prymorsky and Khabarovsk regions of Far East Russia were conducted using a neutralization test. The results revealed that the major cause of HFRS varied with location in Far East Russia: SEOV for Vladivostok city in the Prymorsky region, AMRV in rural areas of the Primorsky region, and probably HTNV for the Khabarovsk region.
Introduction
Hantaviruses are the causative agents of hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome (HPS) in humans. 1,2 Rodents and Soricomorpha species are natural reservoirs of these viruses, and humans acquire infection by inhaling the excreta of infected animals. Hantaviruses are classified in the genus Hantavirus within the family Bunyaviridae, and they possess a genome composed of three negative-stranded RNA segments. The small (S), medium (M), and large (L) genome segments encode nucleocapsid protein (N), two glycoproteins (Gn and Gc), and RNA polymerase, respectively. 3 Each rodent-borne hantavirus has its own host, and more than 40 species of hantaviruses have been identified. 4,5 Some rodent-borne hantaviruses cause HFRS or HPS and are considered to be important zoonotic agents in various countries around the world.
About 20,000–50,000 cases of HFRS are reported annually worldwide, and large proportions of the infections are from the East Eurasian continent, including China, Korea, and Far East Russia. 6–8 Hantaan virus (HTNV) and Seoul virus (SEOV) are known to cause severe and mild forms of HFRS in the Eastern Eurasian Continent, respectively. 2,9,10 HTNV is carried by the striped field mouse, Apodemus agrarius, which preferentially inhabits grass and rice fields. Thus, humans seem to acquire HTNV infection primarily in fields. However, SEOV is carried by the brown rat, Rattus norvegicus, and the black rat, R. rattus, which are peridomestic rodents. Thus, humans tend to become infected with SEOV in cities and close to dwellings. 8,11–14 Recently, Amur virus (AMRV; also known as Soochong virus) was identified in the Korean field mouse A. peninsulae and in HFRS cases in China, Korea, and Far East Russia. 11–13,15,16 Thus, AMRV should also be considered as one of the causative agents of HFRS in East Asia and Far East Russia.
Morbidity of HFRS is 1.9 per 100,000 population, and about 100–200 HFRS cases are reported annually in Far East Russia. 17 In addition, seroprevalence in healthy residents in Khabarovsk and Primorsky regions is about 2.4% and 2.7%, respectively. 18,19 Despite the basic epidemiological information, an etiological analysis of HFRS has not been carried out in this region because of the close antigenicities between the viruses. Usual serological methods, such as indirect immunofluorescent antibody assays, cannot differentiate the infections caused by AMRV and HTNV. Additionally, genetic or antigenic information about the viruses circulating in the region remains extremely limited.
In the present study, we successfully isolated HTNV and AMRV from A. agrarius and A. peninsulae, respectively, in Far East Russia. Furthermore, the genetic and antigenic properties of these isolates were compared with the properties of other hantaviruses. Additionally, a serological differential diagnosis of HFRS patients in the Primorsky and Khabarovsk regions of Far East Russia was performed using a neutralization test to distinguish HTNV, AMRV, and SEOV infections.
Materials and Methods
Rodent survey
Epizootiological surveys targeting rodents were conducted in Khekhtsir, about 20 km south from Khabarovsk, and Galkino, about 20 km east from Khabarovsk, Russia, in 2002 and 2005, respectively. Animals were captured using live traps set in forests and fields at these survey locations. Live animals were killed by cardiac puncture under anesthesia by ether inhalation, and serum was separated from the collected blood. From dead animals, blood samples were collected using filter paper. The paper was air-dried and immersed in a 10× volume of phosphate-buffered saline (PBS) at 4°C overnight, and the supernatant was used as 10× diluted serum. The sera were heat-inactivated at 56°C for 30 minutes and then stored at −40°C until use. The lungs were collected from all rodents and stored at −80°C until use.
Human sera
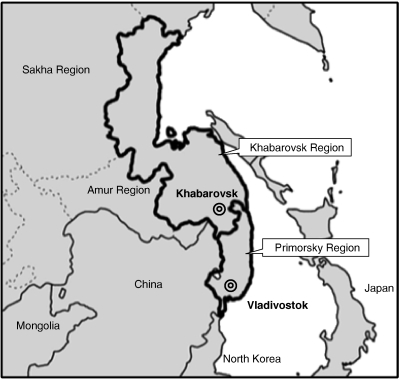
Sera from 67 HFRS patients in the Khabarovsk and Primorsky regions, Russia, from 1965 to 2002 (Figure 1) were tested for neutralizing antibodies to hantaviruses. These sera were heat-inactivated and stored at −40°C until use.
Figure 1.
Geographical locations of epizootiological survey sites in Far East Russia. Rodents were captured in the Khabarovsk region. Sera of HFRS patients were collected in the Primorsky and Khabarovsk regions.
Patient sera were collected with the informed consent of the patients. All experiments using patient sera were approved by the Ethics Committee of Hokkaido University and were performed in a BSL-3 laboratory at the Graduate School of Veterinary Medicine, Hokkaido University.
Cells and media
Vero E6 cells (No. CRL-1586; ATCC, Manassas, VA) were maintained in minimum essential medium with Eagle's salts (MEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 IU/mL penicillin G, and 100 μg/mL streptomycin.
Indirect immunofluorescent antibody assay
For the detection of antihantavirus antibodies in wild rodents, immunofluorescent antibody assay (IFA) was carried out as described previously. 8 Vero E6 cells were infected separately with HTNV 76-118, 20 Bao14, 21 AMRV H5, 22 SEOV SR-11, 23 or PUUV Sotkamo, and the infected cells were fixed using cold acetone on 24-well slides. Wild rodent sera, including sera from dead animals, diluted 1:16 with PBS were spotted onto the slides and incubated at 37°C for 1 hour. The slides were washed three times with PBS, and then, Alexa Fluor 488 goat anti-mouse immunoglobulin G (IgG; Invitrogen) was applied on the slides. After 1 hour of incubation and washing, the cells were observed under a fluorescence microscope. Scattered and fine granular fluorescence in the cytoplasm of Vero E6 cells was considered to indicate a positive reaction. Positive samples were further tested to determine IFA titer, which was the reciprocal of the highest serum dilution giving the positive reaction.
For the detection of hantaviral antigens in Vero E6 cells, the inoculated cells on 24-well slides were visualized using a monoclonal antibody E5/G6 to hantavirus N 24 and Alexa Fluor 488 goat anti-mouse IgG.
Reverse transcription polymerase chain reaction
Total RNA from lung tissue of rodents or Vero E6 cells was extracted using ISOGEN (Nippon Gene Co.). Purified total RNA (5 μg) was reverse-transcribed using Superscript II RNase H reverse transcriptase (Invitrogen) and random primers (Invitrogen) according to the manufacturer's protocol. All segments of hantaviruses were amplified by polymerase chain reaction (PCR) using Platinum Taq DNA polymerase Hi Fidelity (Invitrogen). The thermal conditions for PCR were 94°C for 2 minutes followed by 35 cycles of 94°C for 30 seconds, 50–60°C (depending on the primer used) for 30 seconds, and 68°C for 2 minutes. To detect hantavirus RNA from wild rodents, primers AMR595SFw (5′-agcatgaaggcagaagagat-3′) and Bao14_840SRv (5′-ctgccgtaggtagtccctgt-3′) were used to detect HTNV, and primers AMR595SFw and AMR1252SRv (5′-ctctgtgctagtgttctcaa-3′) were used to detect AMRV.
Virus isolation
The method of hantavirus isolation has been described previously. 25 Lung tissue from both hantavirus antibody- and RNA-positive wild rodents was selected for inoculation in Vero E6 cells. Lungs of A. agrarius and A. peninsulae were homogenized in MEM using a cold pestle, mortar, and sea sand. Part (10%) of the homogenate was centrifuged (2,000 × g for 5 minutes), and each supernatant was inoculated in Vero E6 cells grown in a 25-cm2 flask by centrifugation (670 × g) for 1 hour at room temperature. After the inoculum was discarded, the cells were cultured at 37°C in a 5% CO2 incubator and then subcultured after a 14-day interval. At subculture, some cells were collected and spotted on 24-well slides at 37°C for 4 hours in 5% CO2. The slides were fixed with cold acetone and used as antigen slides for IFA. The remaining collected cells were examined for hantaviral RNA by reverse transcription (RT)-PCR as described above.
Sequencing hantavirus genes
The open reading frames (ORFs) of all segments of newly isolated hantaviruses were amplified using specific primers. PCR products were extracted and purified from agarose gels using the Wizard SV Gel and PCR Clean-Up System (Promega, Fitchburg, WI) and sequenced directly using the BigDye Terminator (version 3.1) Cycle Sequencing Kit and an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
Genetic analysis
Hantavirus nucleotide and amino acid sequences were compared using Genetyx-mac (version 10.0; Genetyx Corp., Tokyo, Japan) for calculating nucleotide and amino acid identities among hantaviruses. MEGA 4 (available at http://www.megasoftware.net/mega.html) was used to generate multiple alignments. A GTR++I model was selected as the best probability model of Bayesian phylogenetic tree of each hantavirus genome segment by MrModeltest software (version 2.3). 26 Phylogenetic trees were drawn using MrBayes (version 3.1.2). 27 Two independent, parallel Markov chain Monte Carlo Metropolis coupling (MCMCMC) analyses, each with four chains (three heated and one cold), were computed for 0.7–1.5 million generations, with tree sampling every 100 generations. A burn-in period of 175,000–375,000 generations was discarded for each run before calculating consensus trees. After two runs of calculation, 10,500–22,500 trees were made, and these trees were used for generating the consensus tree. The sequences of hantaviruses determined in this study were downloaded from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and are listed in Table 1 .
Table 1.
Hantavirus sequences used in this study
| Strain | Source | Capture site | Genbank ID | |||
|---|---|---|---|---|---|---|
| Country | Region | S segment | M segment | L segment | ||
| Newly isolated virus | ||||||
| Khekhtsir/AP209/2005 | A. peninsulae | Russia | Khabarovsk | AB620028 | AB620029 | AB620030 |
| Galkino/AA57/2002 | A. agrarius | Russia | Khabarovsk | AB620031 | AB620032 | AB620033 |
| Amur virus | ||||||
| Solovey/AP61/1999 | A. peninsulae | Russia | Primorsky | AB071183 | –* | – |
| Solovey/AP63/1999 | A. peninsulae | Russia | Primorsky | AB071184 | – | – |
| AP1371 | A. peninsulae | Russia | Khabarovsk | AF427324 | – | – |
| AP1168 | A. peninsulae | Russia | Khabarovsk | AF427323 | – | – |
| AP708 | A. peninsulae | Russia | Khabarovsk | AF427322 | – | – |
| H5 | Human | China | Heilongjiang | AB127996 | AB127993 | |
| B78 | Human | China | Shandong | AB127997 | AB127994 | – |
| Liu | Human | China | Shandong | AF288649 | AF288648 | – |
| JilinAP06 | A. peninsulae | China | Jilin | EF121324 | EF371454 | – |
| SC-1 | A. peninsulae | Korea | Gangwon | AY675349 | AY675353 | DQ056292 |
| SC-2 | A. peninsulae | Korea | Gangwon | AY675350 | DQ056293 | AY675354 |
| Hantaan virus | ||||||
| AA1028 | A. agrarius | Russia | Khabarovsk | AF427318 | – | – |
| AA1719 | A. agrarius | Russia | Khabarovsk | AF427319 | – | – |
| AA2499 | A. agrarius | Russia | Khabarovsk | AF427320 | – | – |
| KHB/HFRS/Pat#A2 | Human | Russia | Khabarovsk | – | AB086620 | – |
| KHB/HFRS/Pat#A3 | Human | Russia | Khabarovsk | – | AB086619 | – |
| KHB/HFRS/Pat#A12 | Human | Russia | Khabarovsk | – | AB086621 | – |
| Bao14 | A. agrarius | China | Heilongjiang | AB127998 | AB127995 | – |
| CGHu1 | Human | China | Guizhou | EU092218 | EU092222 | – |
| S85-46 | A. agrarius | China | Sichuan | AF288659 | AF288658 | – |
| 76-118 | A. agrarius | Korea | Gyong Gi | NC_005218 | NC_005219 | NC_005222 |
| CUMC-B11 | Not known | Korea | U37768 | U38117 | – | |
| CFC94-2 | Human | Korea | X95007 | – | – | |
| Maaji-2 | Human | Korea | AF321095 | – | – | |
| Q32 | A. agrarius | China | Guizhou | AB027097 | DQ371905 | DQ371906 |
| CGAa4MP9 | A. agrarius | China | Guizhou | EF990915 | EF990929 | – |
| 84FLi | A. agrarius | China | Shaanxi | AY017064 | AF345636 | AF336826 |
| A9 | A. agrarius | China | Jiangsu | AF329390 | AF035831 | AF293665 |
| Z10 | Human | China | Zhejiang | NC_006433 | NC_006437 | NC_006435 |
| Seoul virus | ||||||
| SR-11 | Rattus norvegicus | Japan | Hokkaido | M34881 | M34882 | – |
| 80-39 | R. norvegicus | Korea | Seoul | NC_005236 | NC_005237 | NC_005238 |
| Puumala virus | ||||||
| Sotkamo | Myodes glareolus | Finland | NC_005224 | NC_005223 | NC_005225 | |
Not registered or not used in this study.
Immune sera
Antisera to each hantavirus were obtained from hantavirus-immunized mice. Two-week-old BALB/c mice (SLC, Hamamatsu, Japan) were inoculated subcutaneously (s.c.) with 3,000 focus-forming units (FFUs) of two strains of newly isolated hantaviruses. At 70 days post-inoculation (d.p.i.), mice were killed by cardiac puncture under anesthesia with sevoflurane, and the sera were collected. The sera were heat-inactivated and stored at −40°C until use.
All animal experiments were performed according to the Guidelines of Animal Experimentation of the School of Veterinary Medicine, Hokkaido University. All animal experiments were carried out in a biosafety level 3 animal facility.
Focus reduction neutralization test
The protocol for focus reduction neutralization test (FRNT) has been described previously. 28 Immune mouse sera were used to analyze serological relationships among newly isolated hantaviruses and other hantaviruses. Additionally, 67 human sera from HFRS patients in the Primorsky and Khabarovsk regions were used to identify the serotype of hantaviruses infection in each patient. Serially diluted samples (100 μL) were mixed with an equal volume of stock viruses (200 FFUs/100 μL), and these mixtures were incubated at 37°C for 1 hour in 5% CO2. The mixtures (50 μL/well) were inoculated onto Vero E6 cell monolayers grown in 96-well plates (Nalge Nunc, Roskilde, Denmark). After adsorption for 1 hour at 37°C, the inoculum was removed, and 150 μL MEM with 1.5% carboxymethyl cellulose (CMC-MEM) were layered onto the cells and incubated at 37°C for 4 days in 5% CO2. After incubation, the monolayers were washed with PBS, fixed with methanol, and air-dried. Foci of hantaviruses were immunostained with E5/G6 mAb and Alexa Fluor 488 goat anti-mouse IgG. Stained foci were counted under a fluorescence microscope. The neutralization titer was expressed as a reciprocal of the highest dilution that showed ≥ 80% inhibition of virus focus formation.
Statistical analysis
Statistical analysis for the comparison of the proportions of serotypes among HFRS patients living at different sites was performed using the χ 2 test. Haberman's residual analysis was conducted to examine which hantavirus was the major cause of HFRS in the Khabarovsk and Primorsky regions. 29
Results
Rodent survey
We captured 29 A. peninsulae in the forest of Khekhtsir close to Khabarovsk city in 2005 and 47 A. agrarius in the field of Galkino near Khabarovsk city in 2002 (Table 2). Ten A. peninsulae (34.5%) and five A. agrarius (10.6%) were seropositive for hantaviruses. Antibody titers against AMRV in A. peninsulae ranged from 1:32 to 1:1,024, which was fairly comparable with the titers against HTNV. Antibody titers against SEOV were apparently lower than the titers against AMRV in 8 of 10 seropositive A. peninsulae. Antibody titers against HTNV in the five seropositive A. agrarius ranged from 1:1,024 to more than 1:2,048, which was equivalent to the titers against AMRV. Four of five seropositive A. agrarius had IFA antibody titers against AMRV or HTNV that were more than or equal to fourfold greater than the titers against SEOV. No antibody was detected against Puumala virus (PUUV) in any animal. Lung samples of all 76 rodents were tested for the hantavirus S gene by RT-PCR (Table 2). Six A. peninsulae (20.7%) and four A. agrarius (8.5%) contained virus RNA.
Table 2.
Detection of antihantavirus antibodies and virus RNA in A. peninsulae (N = 29) and A. agrarius (N = 47)
| Number | IFA antibody titer* | RT-PCR | Virus isolation | |||
|---|---|---|---|---|---|---|
| AMRV H5 | HTNV 76-118 | SEOV SR-11 | PUUV Sotkamo | |||
| A. peninsulae | ||||||
| 167 | 256 | 512 | < 16 | < 16 | + | N.D.† |
| 172‡ | 256 | 256 | < 16 | < 16 | + | – |
| 186 | 256 | 512 | < 16 | < 16 | + | N.D. |
| 199 | 512 | 128 | < 16 | < 16 | + | N.D. |
| 209 | 1,024 | 1,024 | 128 | < 16 | + | + |
| 161 | 512 | 512 | < 16 | < 16 | – | N.D. |
| 170 | 512 | 128 | 256 | < 16 | – | N.D. |
| 189 | 1,024 | 128 | 128 | < 16 | – | N.D. |
| 190 | 32 | 16 | < 16 | < 16 | – | N.D. |
| 191 | 1,024 | 512 | 512 | < 16 | – | N.D. |
| 166 | < 16 | < 16 | < 16 | < 16 | + | N.D. |
| Others (N = 18) | < 16 | < 16 | < 16 | < 16 | – | N.D. |
| A. agrarius | ||||||
| 56 | > 2,048 | > 2,048 | 128 | < 16 | + | + |
| 57 | > 2,048 | > 2,048 | 512 | < 16 | + | + |
| 61 | 1,024 | 1,024 | 256 | < 16 | + | + |
| 65 | > 2,048 | > 2,048 | > 2,048 | < 16 | + | + |
| 54 | > 2,048 | > 2,048 | 256 | < 16 | – | N.D. |
| Others (N = 42) | < 16 | < 16 | < 16 | < 16 | – | N.D. |
IFA antibody titer was expressed as a reciprocal of the highest dilution that showed specific fluorescence.
N.D. = not done.
Rodent samples in bold were used for virus isolation.
Virus isolation
Virus isolation was carried out using two lung samples of A. peninsulae (numbers 172 and 209) and four samples of A. agrarius (numbers 56, 57, 61, and 65) that were sero- and RNA-positive. Lung homogenates of each sample were inoculated on Vero E6 cells. At 14 d.p.i., hantavirus RNA was detected from cells inoculated with A. peninsulae number 209 and A. agrarius numbers 56, 57, 61, and 65. Hantavirus N was detected at 42 d.p.i. (A. peninsulae number 209) and 28 d.p.i. (A. agrarius numbers 56, 57, 61, and 65). Thus, five strains of hantavirus were isolated from wild rodents (Table 2). Hantavirus isolates from A. peninsulae number 209 and A. agrarius numbers 56, 57, 61, and 65 were designated Khekhtsir/AP209/2005 (AP209), Galkino/AA56/2002 (AA56), Galkino/AA57/2002 (AA57), Galkino/AA61/2002 (AA61), and Galkino/AA65/2002 (AA65), respectively. AP209 and AA57 were used for additional genetic and antigenic characterization.
Genetic analysis of hantavirus isolates
Nucleotide sequences covering the ORF of all segments from AP209 and AA57 were determined. The sequences determined and used in this study are listed in Table 1.
The S segment of AP209 was quite similar to the segment of AMRV Solovey/AP61/1999 and Solovey/AP63/1999 that was detected from A. peninsulae in the Primorsky region, Russia (96.0–96.3% nucleotide identity and 99.3–99.8% amino acid identity) (Table 3) The identities in the S segment between AP209 and AMRVs from North East China and Korea (H5, B78, and SC-1) were slightly lower (90.1–90.5% nucleotide identity and 98.8–99.5% amino acid identity) (Table 3). The nucleotide identities of the S segment between AMRV and other hantaviruses such as HTNV, SEOV, and PUUV were much lower at 82.2–85.8%, 74.1–74.6%, and 63.9–64.3%, respectively. The M and L segments of AP209 were closest to the identities of AMRVs (M segment, 87.5–93.6% and 96.7–99.1%; L segment, 88.6–89.4% and 98.7–99.0% for nucleotide and amino acid identities, respectively) (Table 3) among hantaviruses. Thus, isolate AP209 was identified as AMRV.
Table 3.
Nucleotide (ORF region) and amino acid identities* among the two hantavirus isolates (Khekhtsir AP209 and Galkino AA57) and other hantaviruses
| Strain | AMRV | HTNV | SEOV | PUUV | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Khekhtsir AP209 | Solovey AP61 | Solovey AP63 | H5 | B78 | SC-1 | Galkino AA57 | Bao14 | 76-118 | A9 | Z10 | SR-11 | 80-39 | Sotkamo | |
| S segment | ||||||||||||||
| Khekhtsir AP209 | – | 96.3 | 96.0 | 90.1 | 90.1 | 90.5 | 85.1 | 85.8 | 83.0 | 83.0 | 83.6 | 74.8 | 74.1 | 63.9 |
| Solovey AP61 | 99.8 | – | 99.1 | 90.2 | 90.2 | 90.4 | 84.5 | 85.2 | 82.7 | 82.8 | 83.5 | 73.9 | 74.1 | 64.1 |
| Solovey AP63 | 99.3 | 99.5 | – | 90.1 | 90.1 | 90.4 | 84.2 | 84.9 | 82.8 | 82.6 | 83.4 | 73.8 | 74.0 | 64.3 |
| H5 | 98.8 | 99.1 | 98.6 | – | 100.0 | 91.5 | 83.8 | 83.9 | 83.2 | 82.9 | 83.4 | 74.6 | 73.9 | 64.2 |
| B78 | 98.8 | 99.1 | 98.6 | 100.0 | – | 91.5 | 83.8 | 83.9 | 83.2 | 82.9 | 83.4 | 74.6 | 73.9 | 64.2 |
| SC-1 | 99.5 | 99.8 | 99.3 | 98.8 | 98.8 | – | 83.7 | 84.1 | 83.0 | 84.1 | 84.1 | 74.1 | 73.8 | 64.0 |
| Galkino AA57 | 97.7 | 97.9 | 97.4 | 97.4 | 97.4 | 97.7 | – | 98.4 | 88.8 | 85.5 | 86.4 | 74.3 | 74.7 | 63.5 |
| Bao14 | 97.4 | 97.7 | 97.2 | 97.7 | 97.7 | 97.4 | 99.8 | – | 89.0 | 86.2 | 86.7 | 74.5 | 74.7 | 63.2 |
| 76-118 | 96.5 | 96.7 | 96.3 | 96.7 | 96.7 | 96.5 | 98.8 | 99.1 | – | 84.8 | 85.8 | 74.3 | 74.6 | 63.0 |
| A9 | 96.0 | 96.3 | 95.8 | 96.3 | 96.3 | 96.0 | 97.4 | 97.7 | 96.7 | – | 89.5 | 75.0 | 74.5 | 64.2 |
| Z10 | 97.0 | 97.2 | 96.7 | 97.2 | 97.2 | 97.0 | 97.9 | 98.1 | 97.2 | 97.2 | – | 75.0 | 75.1 | 62.5 |
| SR-11 | 82.1 | 82.1 | 81.6 | 81.6 | 81.6 | 81.8 | 82.3 | 82.1 | 82.3 | 80.9 | 81.1 | – | 97.6 | 62.0 |
| 80-39 | 83.0 | 83.0 | 82.5 | 82.5 | 82.5 | 82.8 | 83.2 | 83.0 | 83.2 | 81.8 | 82.1 | 98.4 | – | 62.0 |
| Sotkamo | 60.5 | 60.5 | 60.3 | 60.7 | 60.7 | 60.3 | 60.5 | 60.7 | 60.7 | 59.8 | 60.0 | 61.7 | 61.9 | – |
| M segment | ||||||||||||||
| Khekhtsir AP209 | – | – | – | 93.6 | 93.1 | 87.5 | 81.2 | 81.2 | 81.1 | 80.3 | 80.1 | 72.3 | 71.8 | 59.3 |
| H5 | 98.9 | – | – | – | 96.5 | 87.0 | 79.9 | 79.9 | 79.8 | 79.4 | 79.8 | 71.7 | 71.2 | 60.1 |
| B78 | 98.7 | – | – | 99.1 | – | 87.0 | 80.4 | 80.4 | 79.6 | 79.7 | 79.5 | 71.6 | 71.3 | 59.9 |
| SC-1 | 96.7 | – | – | 96.9 | 97.1 | – | 80.6 | 80.3 | 80.5 | 79.4 | 80.0 | 72.2 | 71.7 | 59.4 |
| Galkino AA57 | 92.4 | – | – | 92.7 | 92.3 | 91.7 | – | 97.0 | 87.6 | 84.0 | 85.0 | 72.0 | 71.8 | 60.1 |
| Bao14 | 92.3 | – | – | 92.5 | 92.2 | 91.5 | 99.0 | – | 87.7 | 84.4 | 85.1 | 72.3 | 71.9 | 59.8 |
| 76-118 | 92.3 | – | – | 92.2 | 92.0 | 91.1 | 97.5 | 97.9 | – | 84.6 | 84.3 | 72.5 | 72.0 | 60.1 |
| A9 | 91.6 | – | – | 91.5 | 91.2 | 90.6 | 94.7 | 95.0 | 95.4 | – | 87.3 | 71.8 | 71.6 | 60.4 |
| Z10 | 92.0 | – | – | 91.9 | 91.5 | 90.7 | 94.9 | 95.0 | 95.3 | 96.2 | – | 71.6 | 71.6 | 60.0 |
| SR-11 | 76.5 | – | – | 76.8 | 76.3 | 76.4 | 76.6 | 76.8 | 77.0 | 76.3 | 76.4 | – | 96.5 | 60.8 |
| 80-39 | 76.8 | – | – | 77.0 | 76.4 | 76.6 | 76.8 | 77.0 | 77.1 | 76.3 | 76.4 | 98.9 | – | 60.7 |
| Sotkamo | 54.3 | – | – | 54.5 | 54.4 | 53.9 | 53.8 | 53.6 | 53.8 | 53.6 | 53.7 | 53.8 | 53.8 | – |
| L segment | – | – | ||||||||||||
| Khekhtsir AP209 | – | – | – | 89.4 | – | 88.9 | 81.9 | 81.9 | 81.5 | 81.6 | 82.2 | – | 75.6 | 67.5 |
| H5 | 99.0 | – | – | – | – | 88.6 | 81.6 | 81.2 | 81.7 | 81.0 | 81.8 | – | 75.4 | 67.7 |
| SC-1 | 98.7 | – | – | 98.7 | – | – | 95.9 | 81.1 | 81.3 | 81.1 | 81.4 | – | 75.3 | 67.4 |
| Galkino AA57 | 96.1 | – | – | 96.0 | – | 81.2 | – | 97.3 | 88.0 | 83.4 | 84.0 | – | 74.9 | 67.3 |
| Ba014 | 96.1 | – | – | 95.9 | – | 95.8 | 99.7 | – | 88.1 | 83.2 | 84.0 | – | 74.8 | 67.2 |
| 76-118 | 95.7 | – | – | 95.8 | – | 95.6 | 98.7 | 98.6 | – | 83.6 | 83.5 | – | 74.3 | 66.8 |
| A9 | 94.2 | – | – | 94.3 | – | 93.9 | 95.8 | 95.6 | 95.6 | – | 85.4 | – | 74.7 | 67.1 |
| Z10 | 95.5 | – | – | 95.5 | – | 95.3 | 97.6 | 97.5 | 97.3 | 96.1 | – | – | 74.9 | 67.3 |
| 80-39 | 85.5 | – | – | 85.3 | – | 85.3 | 85.4 | 85.4 | 85.0 | 83.7 | 85.4 | – | – | 67.7 |
| Sotkamo | 69.1 | – | – | 68.7 | – | 68.7 | 69.1 | 69.0 | 68.9 | 67.9 | 69.3 | – | 68.5 | – |
Values to the right above the diagonal show nucleotide identities; values to the left below the diagonal show amino acid identities.
The nucleotide and amino acid sequences of all genome segments of AA57 were quite similar to the sequences of HTNV strain Bao14, which was isolated from an HFRS patient in Heilongjiang, China (S segment, 98.4% and 99.8%; M segment, 97.0% and 99.0%; L segment, 97.3% and 99.7% nucleotide and amino acid identities, respectively) (Table 3). Thus, isolate AA57 was identified as HTNV. Additionally, a comparison of the M segments between AA57 and hantavirus sequences from HFRS patients in the Khabarovsk region (KHB/HFRS/PAT#A2, KHB/HFRS/PAT#A3, and KHB/HFRS/PAT#A12) 12 showed 96–99% identity at the nucleotide level. The nucleotide identities of the S, M, and L segments between AA57 and other HTNVs (76-118, A9, and Z10) ranged from 84.8% to 98.4%, from 84.0% to 97.0%, and from 84.0% to 97.3%, respectively (Table 3).
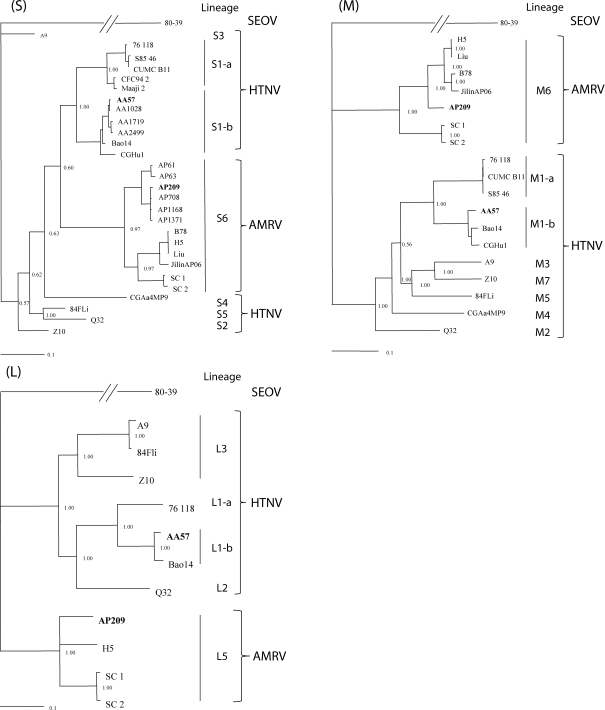
Phylogenetic analyses of hantavirus S, M, and L segments were performed (Figure 2). Previously, the work by Zou and others 30 described that the Hantaan superclade, including HTNV and AMRV, was divided into several lineages, and the S, M, and L segments of AMRV occupied lineages S6, M6, and L5, respectively. In the present study, the S, M, and L segments of AP209 clustered with AMRVs in lineages S6, M6, and L5, respectively, consistent with the results in the work by Zou and others. 30 AP209 was more related to AMRVs detected in the Primorsky region of Far East Russia, such as AP708, AP1371, AP1168, Solovey/AP61/1999, and Solovey/AP63/1999, than AMRVs from China and Korea. Although the M and L segments of AMRV clustered in distinct lineages from other HTNVs, the S segment of AMRV clustered within HTNV. The S, M, and L segments of strain AA57 clustered with HTNVs from the Primorsky region (AA1028, AA1719, and AA2499) and HTNVs from China (Bao14 and CGHu1), which consist of the Far East (FE) genotype within HTNVs. 13, 28 Interestingly, strain CGHu1 isolated from Guizhou, a southern province of China, also clustered with the FE genotype. HTNV lineages in S1 were divided into S1-a and S1-b sublineages according to their geographical origins: S1-a from Korea (76-118, CUMC-B11, and Maaji-2) and S1-b from Far East Russia (AA57, AA1028, AA1719, and AA2499) and China (Bao14 and CGHu1). The same clustering patterns were also observed with the M and L segments.
Figure 2.
Phylogenetic consensus trees of hantaviruses based on nucleotide sequences covering the entire ORF of the S, M, and L segments. The trees were generated using MCMCMC analyses. The reliability of the tree was evaluated by the posterior probability value derived from MrBayes. The scale bar indicates 0.1 nucleotide substitutions per site.
Antigenic characterization
The antigenic characterization of AP209, AA57, and other hantaviruses was performed using a neutralization test (Table 4). Anti-AMRV mouse sera had more than fourfold higher titers against AMRV strains H5 and AP209 than those titers against HTNV and SEOV. Anti-AA57 mouse serum had a more than fourfold higher neutralization titer against AA57 compared with those titers against AMRVs and SEOV. These results indicate that the antigenicities of AMRV, HTNV, and SEOV are different and that the neutralization test may be useful in the differential diagnosis of HFRS caused by AMRV, HTNV, or SEOV in Far East Russia.
Table 4.
Antigenic characterization of Khekhtsir AP209 and Galkino AA57 by cross-neutralization test using immune mouse sera
| Antisera | Neutralization titer* | ||||
|---|---|---|---|---|---|
| AMRV | HTNV | SEOV | |||
| Virus | Strain | H5 | Khekhtsir AP209 | Galkino AA57 | SR-11 |
| AMRV | H5 | 80 | 80 | 20 | < 20 |
| AMRV | Khekhtsir AP209 | 320 | 640 | 40 | 20 |
| HTNV | Galkino AA57 | 640 | 640 | 2,560 | 40 |
| SEOV | SR-11 | < 20 | < 20 | < 20 | 40 |
Neutralization titer was expressed as a reciprocal of the highest dilution that showed 80% or more inhibition of virus focus formation.
Serological analysis of patient sera
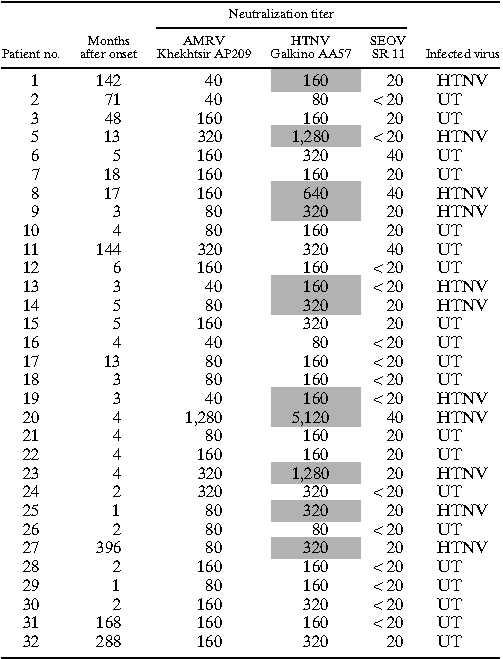
To examine the causative agents of HFRS in the Khabarovsk and Primorsky regions, the neutralization test was performed on a total of 67 HFRS patients who had IFA antibodies to hantaviruses (Tables 5 and 6). The hantavirus in each HFRS patient was determined as a certain virus giving a neutralization titer more than or equal to fourfold higher than the titer of other viruses (Tables 5 and 6). If the neutralization titer difference was less than fourfold, we considered the infected hantavirus of the sample to be untyped. If the neutralization titer was less than 1:20 for any hantaviruses, we regarded the sample as negative for hantavirus infection. Of 67 HFRS patients, 14, 13, and 15 were diagnosed as AMRV (20.9%), HTNV (19.4%), and SEOV (22.4%) infections, respectively, and 25 patients (37.3%) were untyped (Tables 5 and 6). All of 20 untyped sera in Khabarovsk region showed strong cross-reaction to AMRV and HTNV (Table 6). The seroprevalence of AMRV, HTNV, and SEOV in HFRS patients differed with each sampling site (χ2 value = 70.69, degrees of freedom [df] = 6, P < 0.001). SEOV (60.0%; P < 0.001) and AMRV (81.8%; P < 0.001) were the major causative agents of HFRS in Vladivostok city and the rural areas of Primorsky region except Vladivostok, respectively (Table 5). HTNV (35.5%; P < 0.002) may be one of the major causes in the Khabarovsk region (Table 6).
Table 5.
Differential diagnosis of HFRS patients in the Primorsky region by neutralization test
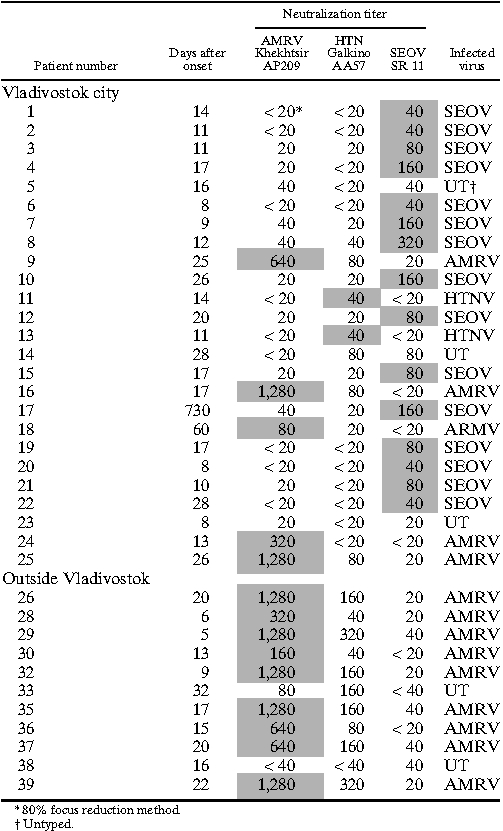
Table 6.
Differential diagnosis of HFRS patients in the Khabarovsk region by neutralization test
Discussion
Previous investigations of HFRS patients in Far East Russia have revealed that AMRV, HTNV, and SEOV are causative agents of HFRS in this region. 12,13 However, no complete etiological analysis of each HFRS patient to identify the infected hantavirus has been conducted. Because the biological properties of hantaviruses in this region have not been well-characterized, a differential diagnosis of infected viruses in HFRS is quite difficult. Thus, we isolated hantaviruses in Far East Russia and characterized the new isolates. Additionally, serological differential diagnoses were carried out on HFRS patients in this region to identify the virus in each infected patient.
Rodent surveys were carried out in rural areas of the Khabarovsk region, and we confirmed that HTNV and AMRV were maintained in A. agrarius and A. peninsulae, respectively, which we have described previously for hantavirus infection in rodents of the Primorsky region, Russia. 8,11 These results indicated that HTNV and AMRV are distributed throughout Far East Russia.
We successfully isolated four strains of HTNV and one strain of AMRV. Among the isolates, HTNV strain AA57 from A. agrarius and AMRV strain AP209 were subjected to genetic and antigenic characterization. AA57 was genetically closely related to the sequences from HFRS patients in the Khabarovsk region 12 and Bao14, which was isolated from HFRS patients in China. Newly isolated AMRV AP209 from A. peninsulae was genetically closely related to the sequences from rodents and HFRS patients in the Primorsky region. These results indicate that HTNV and AMRV are the causative agents of HFRS in the Khabarovsk region. Although the M and L segments of AMRV and HTNV clustered in distinct lineages, the S segment of AMRV was included in HTNV. This observation was also reported in the work by Zou and others. 30 Previous reports of phylogenetic analysis have indicated that a natural reassortment occurred between HTNV and SEOV in China and among different lineages of PUUVs in nature. 31,32 Additionally, dual infection with Sin Nombre virus and Black Creek Canal virus could produce reassortant viruses under experimental conditions. 33 Other reports have described that a recombination of the virus genome occurred in Dobrava virus and Saaremaa virus and in different lineages of Tula viruses in nature. 34,35 Thus, it is feasible that some discontinuous mutation, such as reassortment, may have occurred between AMRV and HTNV as indicated between HTNV and SEOV in China. 32
In the cross-neutralization test using immune sera against HTNV, AMRV, and SEOV, the immune sera had fourfold higher titers against the homologous virus than the heterologous viruses. This result indicates that the cross-neutralization test is useful for the differential diagnosis of AMRV, HTNV, and SEOV infection in Far East Russia.
To obtain more information about HFRS etiology and epidemiology in Far East Russia, the neutralization tests were carried out using HFRS patient sera from the Primorsky and Khabarovsk regions. The results showed that AMRV (20.9%), HTNV (19.4%), and SEOV (22.4%) were the causative agents of HFRS in Far East Russia, and these findings are consistent with the results of previous reports. 13 However, the major cause of HFRS varied according to location. SEOV was the most common cause of HFRS in Vladivostok city in the Primorsky region (15/25). In Vladivostok city, R. norvegicus and R. rattus may transmit SEOV to city residents (Table 5). Some patients in Vladivostok were infected with AMRV and HTNV (5/25 and 2/25, respectively). Russian people frequently visit vegetable gardens in rural areas called dachas to cultivate plants. Because A. peninsulae and A. agrarius may live in forests and fields near dachas, it is highly possible that people, including city residents, may acquire AMRV or HTNV infection close to dachas. Interestingly, in the rural area of the Primorsky region, most patients were infected with AMRV (9/11) (Table 5). It is possible that people in this area may become infected more frequently with AMRV in forests that A. peninsulae inhabit. However, in the Khabarovsk region, HTNV may be the major cause of HFRS (Table 6). AMRV infection may occur in the Khabarovsk region, although no AMRV-specific antibodies were detected in this study. There are three possible explanations for negative results of AMRV-specific antibodies. (1) Residents in Khabarovsk region may contact more frequently with A. agrarius than A. peninsulae. (2) Because of the similar antigenicity between AMRV and HTNV, AMRV-infected patients may produce antibodies cross-reacted to HTNV. Most patient sera from the Khabarovsk region were collected in late convalescent phase (Table 6), which may have influenced the results of neutralization test. (3) The possibility of dual infections with AMRV and HTNV cannot be ruled out. However, because A. agrarius and A. peninsulae preferentially inhibit grass field and forest, respectively, the possibility of the dual infection at the same time may be extremely low. In addition, consecutive infections of the two viruses may not occur, because patients infected with AMRV or HTNV can produce cross-neutralizing antibodies to the homologous and heterologous viruses. Additional studies should be conducted to develop a clear picture of HFRS epidemiology on a larger scale. Unfortunately, in some patients (25/71), we could not differentiate the infecting hantavirus. Serotype-specific conformational epitopes have been detected in the center and C-terminal regions of the N. 24,36 Thus, a serotyping system using N-terminal truncated N could be a solution for the differential diagnosis of hantavirus infections. 37,38
In conclusion, new isolates of AMRV and HTNV were characterized, and AMRV, HTNV, and SEOV infections were differentiated in HFRS patients of Far East Russia. The major cause of HFRS varied by location in the region: SEOV for Vladivostok city in the Prymorsky region, AMRV in the rural areas of the Primorsky region, and probably HTNV for the Khabarovsk region. Additional study is necessary to determine the risk factors of HFRS for each pathogenic virus in Far East Russia.
ACKNOWLEDGMENTS
The authors thank Dr. Vladimir A. Demenev for arranging the sampling of HFRS patient sera. We appreciate all of the people who worked with us in the field in the Khabarovsk region helping with rodent trapping and supporting the surveys.
Footnotes
Financial support: This study was supported financially by grants-in-aid for scientific research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a scientific grant for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Welfare, and Labour of Japan, and a grant from the Program of Global Center of Excellence Program, Hokkaido University (“Establishment of International Collaboration Centers for Zoonosis Control”).
Authors’ addresses: Hiroaki Kariwa, Keisuke Yoshikawa, Yoichi Tanikawa, Takahiro Seto, Takahiro Sanada, Ngonda Saasa, Kentaro Yoshii, and Ikuo Takashima, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan, E-mails: kariwa@vetmed.hokudai.ac.jp, canis-familiaris-l@k.vodafone.ne.jp, tanikawa.yf@om.asahi-kasei.co.jp, setotaka@vetmed.hokudai.ac.jp, sanada-t@vetmed.hokudai.ac.jp, nsaasa@yahoo.co.uk, kyoshii@vetmed.hokudai.ac.jp, and takasima@vetmed.hokudai.ac.jp. Leonid I. Ivanov, Plague Control Station of Khabarovsk, Khabarovsk, Russia, E-mail: chum@chum.khv.ru. Raisa Slonova, Research Institute of Epidemiology and Microbiology, Siberian Branch of Russian Academy of Medical Sciences, Vladivostok, Russia, E-mail: atavalk@inbox.ru. Tatyana A. Zakharycheva, Far Eastern State Medical University, Khabarovsk, Russia, E-mail: dolika@inbox.ru. Ichiro Nakamura, Research Center for Zoonosis Control, Hokkaido University, Sapporo, Japan, E-mail: inaka@czc.hokudai.ac.jp. Kumiko Yoshimatsu and Jiro Arikawa, Graduate School of Medicine, Hokkaido University, Sapporo, Japan, E-mails: yosimatu@med.hokudai.ac.jp and j_arika@med.hokudai.ac.jp.
References
- 1.Krüger DH, Ulrich R, Lundkvist A. Hantavirus infections and their prevention. Microbes Infect. 3: 2001:1129–1144. doi: 10.1016/s1286-4579(01)01474-5. [DOI] [PubMed] [Google Scholar]
- 2.Schmaljohn CS, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonsson CB, Schmaljohn CS. In: Hantavirus. Berlin. Schmaojohn CS, Nichol ST, editors. Germany: Springer-Verlag; 2001. pp. 15–32. (Replication of hantavirus). [Google Scholar]
- 4.Jonsson CB, Figueiredo LTM, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliot RM, Beaty BJ, Calisher CH, Goldbach RW, Nichol ST, Plyusnin A, Schmaljohn CS. In: Virus Taxonomy. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. London, United Kingdom: Elsevier Academic Press; 2005. pp. 695–715. (Family Bunyaviridae). [Google Scholar]
- 6.Zhang YZ, Zou Y, Fu ZF, Plyusnin A. Hantavirus infections in humans and animals, China. Emerg Infect Dis. 2010;16:1195–1203. doi: 10.3201/eid1608.090470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song JY, Chun BC, Kim SD, Baek LJ, Kim SH, Sohn JW, Cheong HJ, Kim WJ, Park SC, Kim MJ. Epidemiology of hemorrhagic fever with renal syndrome in endemic area of the Republic of Korea, 1995–1998. J Korean Med Sci. 2006;21:614–620. doi: 10.3346/jkms.2006.21.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kariwa H, Lokugamage K, Lokugamage N, Miyamoto H, Yoshii K, Nakauchi M, Yoshimatsu K, Arikawa J, Ivanov LI, Iwasaki T, Takashima I. A comparative epidemiological study of hantavirus infection in Japan and far east Russia. Jpn J Vet Res. 2007;54:145–161. [PubMed] [Google Scholar]
- 9.Kariwa H, Zhong CB, Araki K, Yoshimatsu K, Lokugamage K, Lokugamage N, Murphy ME, Mizutani T, Arikawa J, Fukushima H, Xiong H, Jiehua C, Takashima I. Epizootiological survey of hantavirus among rodent species in Ningxia Hui Autonomous Province, China. Jpn J Vet Res. 2001;49:105–114. [PubMed] [Google Scholar]
- 10.Song G, Hang CS, Liao HX, Fu JL, Gao GZ, Qiu HL, Zhang QF. Antigenic difference between viral strains causing classical and mild types of epidemic hemorrhagic fever with renal syndrome in China. J Infect Dis. 1984;150:889–894. doi: 10.1093/infdis/150.6.889. [DOI] [PubMed] [Google Scholar]
- 11.Lokugamage K, Kariwa H, Hayasaka D, Cui BZ, Iwasaki T, Lokugamage N, Ivanov LI, Volkov VI, Demenev VA, Slonova R, Kompanets G, Kushnaryova T, Kurata T, Maeda K, Araki K, Mizutani T, Yoshimatsu K, Arikawa J, Takashima I. Genetic characterization of hantaviruses transmitted by the Korean field mouse (Apodemus peninsulae), far east Russia. Emerg Infect Dis. 2002;8:768–776. doi: 10.3201/eid0808.010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamoto H, Kariwa H, Araki K, Lokugamage K, Hayasaka D, Cui BZ, Lokugamage N, Ivanov LI, Mizutani T, Iwasa MA, Yoshimatsu K, Arikawa J, Takashima I. Serological analysis of hemorrhagic fever with renal syndrome (HFRS) patients in far eastern Russia and identification of the causative hantavirus genotype. Arch Virol. 2003;148:1543–1556. doi: 10.1007/s00705-003-0113-x. [DOI] [PubMed] [Google Scholar]
- 13.Yashina LN, Patrushev NA, Ivanov LI, Slonova RA, Mishin VP, Kompanez GG, Zdanovskaya NI, Kuzina II, Safronov PF, Chizhikov VE, Schmaljohn C, Netesov SV. Genetic diversity of hantaviruses associated with hemorrhagic fever with renal syndrome in the far east of Russia. Virus Res. 2000;70:31–44. doi: 10.1016/s0168-1702(00)00203-3. [DOI] [PubMed] [Google Scholar]
- 14.Yashina L, Mishin V, Zdanovskaya N, Schmaljohn C, Ivanov L. A newly discovered variant of a hantavirus in Apodemus peninsulae, far eastern Russia. Emerg Infect Dis. 2001;7:912–913. doi: 10.3201/eid0705.017530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lokugamage K, Kariwa H, Lokugamage N, Iwasa M, Hagiya T, Araki K, Tachi A, Mizutani T, Yoshimatsu K, Arikawa J, Iwasaki T, Takashima I. Comparison of virulence of various hantaviruses related to hemorrhagic fever with renal syndrome in newborn mouse model. Jpn J Vet Res. 2004;51:143–149. [PubMed] [Google Scholar]
- 16.Baek LJ, Kariwa H, Lokugamage K, Yoshimatsu K, Arikawa J, Takashima I, Kang JI, Moon SS, Chung SY, Kim EJ, Kang HJ, Song KJ, Klein TA, Yanagihara R, Song JW. Soochong virus: an antigenically and genetically distinct hantavirus isolated from Apodemus peninsulae in Korea. J Med Virol. 2006;78:290–297. doi: 10.1002/jmv.20538. [DOI] [PubMed] [Google Scholar]
- 17.Garanina SB, Platonov AE, Zhuravlev VI, Murashkina AN, Yakimenko VV, Korneev AG, Shipulin GA. Genetic diversity and geographic distribution of hantaviruses in Russia. Zoonoses Public Health. 2009;56:297–309. doi: 10.1111/j.1863-2378.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov LI, Liberova RN, Zdanovskaya NI, Denisov EA, Prosina NA. Contingents of people for specific prevention of hemorrhagic fever with renal syndrome in the Khabarovsk Territory and Jewish Autonomous Region of Russia. Far East Med J 4 (Suppl Vaccination): 1996:44–48. [Google Scholar]
- 19.Kompanets GG, Slonova RA, Kushnareva TV, Maksema IG, Pysina TV, Kraeva LS. Humoral immune response to hantaviruses in health people of the Maritime Territory of Russia. Zh Microbiol (Moscow) 2006;3((Suppl)):84–87. [Google Scholar]
- 20.Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean Hemorrhagic fever. J Infect Dis. 1978;137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Yoshimatsu K, Ebihara H, Ogino M, Araki K, Kariwa H, Wang Z, Luo Z, Li D, Hang C, Arikawa J. Genetic diversity of hantaviruses isolated in china and characterization of novel hantaviruses isolated from Niviventer confucianus and Rattus rattus. Virology. 2000;278:332–345. doi: 10.1006/viro.2000.0630. [DOI] [PubMed] [Google Scholar]
- 22.Liang M, Li D, Xiao SY, Hang C, Rossi CA, Schmaljohn CS. Antigenic and molecular characterization of hantavirus isolates from China. Virus Res. 1994;31:219–233. doi: 10.1016/0168-1702(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura T, Morita C, Komatsu T, Sugiyama K, Arikawa J, Shiga S, Takeda H, Akao Y, Imaizumi K, Oya A, Hashimoto N, Urasawa S. Isolation of virus causing hemorrhagic fever with renal syndrome (HFRS) through a cell culture system. Jpn J Med Sci Biol. 1983;36:17–25. doi: 10.7883/yoken1952.36.17. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimatsu K, Arikawa J, Tamura M, Yoshida R, Lundkvist A, Niklasson B, Kariwa H, Azuma I. Characterization of the nucleocapsid protein of Hantaan virus strain 76-118 using monoclonal antibodies. J Gen Virol. 1996;77:695–704. doi: 10.1099/0022-1317-77-4-695. [DOI] [PubMed] [Google Scholar]
- 25.Kariwa H, Arikawa J, Takashima I, Isegawa Y, Yamanishi K, Hashimoto N. Enhancement of infectivity of hantavirus in cell culture by centrifugation. J Virol Methods. 1994;49:235–244. doi: 10.1016/0166-0934(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 26.Nylander JAA. MrModeltest v2. 2004. http://www.abc.se/~nylander/mrmodeltest2/mrmodeltest2.html Available at. Accessed May 22, 2008. [Google Scholar]
- 27.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 28.Lokugamage K, Kariwa H, Lokugamage N, Miyamoto H, Iwasa M, Hagiya T, Araki K, Tachi A, Mizutani T, Yoshimatsu K, Arikawa J, Takashima I. Genetic and antigenic characterization of the Amur virus associated with hemorrhagic fever with renal syndrome. Virus Res. 2004;101:127–134. doi: 10.1016/j.virusres.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Shelby JH. The analysis of residuals in cross classified tables. Biometrics. 1973;29:205–220. [Google Scholar]
- 30.Zou Y, Hu J, Wang ZX, Wang DM, Li MH, Ren GD, Duan ZX, Fu ZF, Plyusnin A, Zhang YZ. Molecular diversity and phylogeny of Hantaan virus in Guizhou, China: evidence for Guizhou as a radiation center of the present Hantaan virus. J Gen Virol. 2008;89:1987–1997. doi: 10.1099/vir.0.2008/000497-0. [DOI] [PubMed] [Google Scholar]
- 31.Razzauti M, Plyusnina A, Henttonen H, Plyusnin A. Accumulation of point mutations and reassortment of genomic RNA segments are involved in the microevolution of Puumala hantavirus in a bank vole (Myodes glareolus) population. J Gen Virol. 2008;89:1649–1660. doi: 10.1099/vir.0.2008/001248-0. [DOI] [PubMed] [Google Scholar]
- 32.Zou Y, Hu J, Wang ZX, Wang DM, Yu C, Zhou JZ, Fu ZF, Zhang YZ. Genetic characterization of hantaviruses isolated from Guizhou, China: evidence for spillover and reassortment in nature. J Med Virol. 2008;80:1033–1041. doi: 10.1002/jmv.21149. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez LL, Owens JH, Peters CJ, Nichol ST. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. 1998;242:99–106. doi: 10.1006/viro.1997.8990. [DOI] [PubMed] [Google Scholar]
- 34.Klempa B, Schmidt HA, Ulrich R, Kaluz S, Labuda M, Meisel H, Hjelle B, Krüger DH. Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J Virol. 2003;77:804–809. doi: 10.1128/JVI.77.1.804-809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibold C, Meisel H, Krüger DH, Labuda M, Lysy J, Kozuch O, Pejcoch M, Vaheri A, Plyusnin A. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruo SL, Sanchez A, Elliott LH, Brammer LS, McCormick JB, Fisher-Hoch SP. Monoclonal antibodies to three strains of hantaviruses: Hantaan, R22, and Puumala. Arch Virol. 1991;119:1–11. doi: 10.1007/BF01314318. [DOI] [PubMed] [Google Scholar]
- 37.Araki K, Yoshimatsu K, Ogino M, Ebihara H, Lundkvist A, Kariwa H, Takashima I, Arikawa J. Truncated hantavirus nucleocapsid proteins for serotyping Hantaan, Seoul, and Dobrava hantavirus infections. J Clin Microbiol. 2001;39:2397–2404. doi: 10.1128/JCM.39.7.2397-2404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura I, Yoshimatsu K, Lee BH, Okumura M, Taruishi M, Araki K, Kariwa H, Takashima I, Arikawa J. Development of a serotyping ELISA system for Thailand virus infection. Arch Virol. 2008;153:1537–1542. doi: 10.1007/s00705-008-0128-4. [DOI] [PubMed] [Google Scholar]