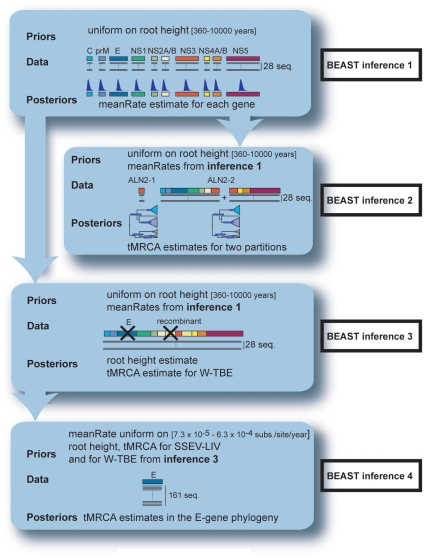
Figure 1. Analytical framework for the BEAST analyses.
Inference 1 analyses the variation in substitution rates across the genome for the 28 full length ORFs. The inferred posterior probability distributions of meanRates for individual genes are set as priors for inferences 2 and 3. Inference 2 dates the recombination event based on the 28 full length ORFs. Dates are evaluated separately for the recombinant region and for the non-recombinant sequences. Inference 3 gathers priors information for root height, tMRCA(W-TBE) and tMRCA(LIV & SSEV) used for the next inference. The alignment for inference 3contains the same 28 sequences, with the deletion of E-gene and the recombinant region. Inference 4 refines estimates for tMRCA(W-TBEV) based on 161 E-sequences. Prior distribution for meanRate parameter is derived from the literature. Priors for tMRCA(W-TBEV) and tMRCA(LIV & SSEV) were obtained from BEAST inference 3.