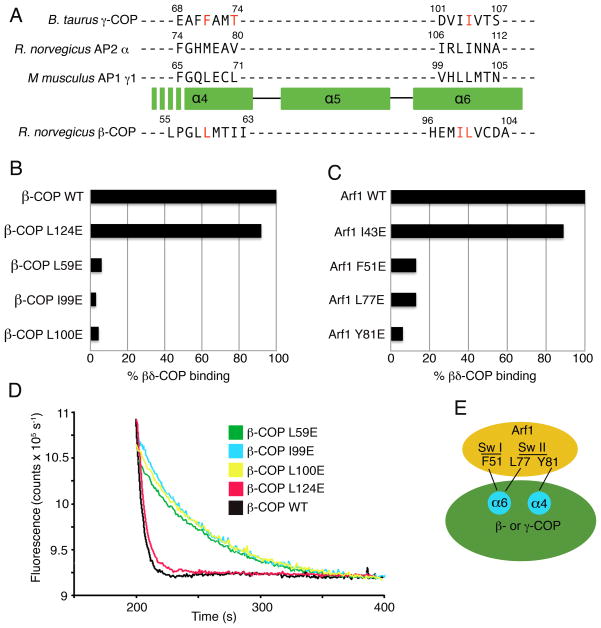
Figure 5. Mapping the Arf1-binding site on βδ-COP.
(A) Sequence- and structure-based alignment of the large subunits of COPI and AP complexes, in the vicinity of the α4- and α6-helices of the α-solenoid domain. The three upper sequences are aligned based on a structural overlap of γ-COP (this study), α2-adaptin (from the crystal structure of AP2 determined by Collins et al. [2002]) and γ1-adaptin (from the AP1 crystal structure determined by Heldwein et al. [2004]). The sequence of β-COP is aligned to the others based on sequence homology including the conservation of helical repeats of the α-solenoid fold. The residues of γ-COP and β-COP selected for mutagenesis are colored red. The location of β-COP residue Pro56 implies that the α4 helix will start downstream of this residue in β-COP; consistent with this, the key interface residues of γ-COP are located toward the C-terminal end of the α4 helix.
(B) Bar graph shows the effects of β-COP mutations on the interaction between βδ-COP and Arf1, as measured by the pull-down assay. Mutations were introduced into full-length β-COP protein. The β-COP residue L124 was selected as a site distant from the interface with Arf1; this residue corresponds to the mutation L128E on γ-COP (see Figure 4A).
(C) Bar graph shows the effects of mutations in S. cerevisiae Arf1 on the interaction with full-length βδ-COP. This is the same set of mutations that were used to test γζ-COP interactions in Figure 4D.
(D) The ability of β-COP mutants to synergize with GAP in GTP hydrolysis was measured using the fluorescence assay. The effect of the β-COP mutations is essentially the same as in the pull-down assay (B). Curves have been offset incrementally along the x-axis.
(E) Summary of the mapping experiments. Arf1 probably binds in a similar manner to γ-COP and β-COP, whereby the Arf1 switch regions interact with residues of the α4- and α6-helices of the α-solenoid domain.