FIGURE 3.
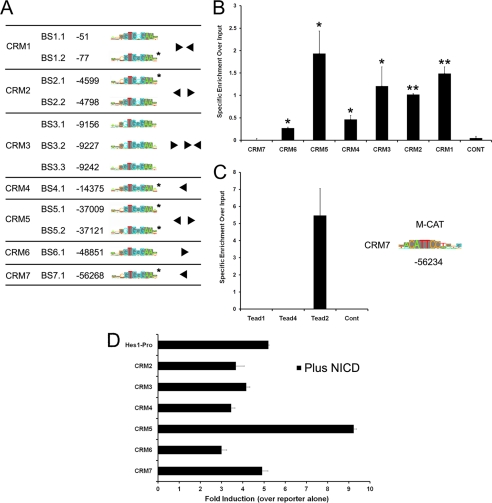
hes1 CRMs are bound by the RBP-JK and TEAD2 transcription factors. A, identification of RBP-JK motifs in the hes1 CRMs using the BiFa discovery tool. The position of each RBP-JK motif relative to the hes1 TSS is shown. Sequences matching the consensus high affinity PSSM are marked in gray and the architecture of the RPB-JK BSs within each CRM is indicated. Conserved BSs are marked by an asterisk. B, RBP-JK binds to hes1 CRMs in proliferating C2C12 cells. ChIP assays were performed using either an antibody against RBP-JK or an isotype-specific anti-GFP control (n = 4). The precipitated DNA fragments were PCR-amplified using primers spanning the predicted RBP-JK BSs within each CRM. The control represents an intervening region in the hes1 upstream sequence in which no RBP-JK BSs were detected in silico. Analysis using an unpaired Student's t test shows that RBP-JK occupancy at CRM1–6 is statistically significant compared with a nonbinding control region. * indicates p < 0.05; ** indicates p < 0.01. C, position of the conserved M-CAT motif in hes1 CRM7 and sequence matches to the consensus PSSM are shown. ChIP assays were performed in proliferating C2C12 cells using anti-TEAD1, anti-TEAD2, anti-TEAD4, or anti-GFP (isotype control) antibodies. Precipitated DNA fragments were PCR-amplified using primers spanning the predicted M-CAT motif in CRM7. To calculate specific enrichment over input, the signal for each PCR was quantified and divided by the input, and the background intensity, measured using an IgG isotope control, was subtracted. D, modulation of hes1 CRM-pro reporter activity by active NOTCH. Hes1CRM-reporter constructs were co-transfected into C2C12 cells along with a 1:1 ratio of reporter to NICD overexpression vector or an empty control vector to make the total amount of DNA transfected in each case equal. The results are presented as fold induction relative to the activity of each reporter in the absence of NICD and represent a mean ± S.D. of three independent experiments.