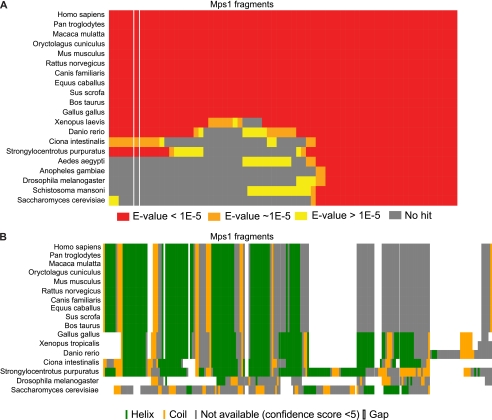
FIGURE 3.
Mps1 TPR domain appeared after the emergence of deuterostomes. A, human Mps1 overlapping fragments (length, 160 amino acids; offset, 10 amino acids) were blasted (PSI-BLAST) against the entire NCBI sequence database (nonredundant). For each listed organism for which we could find a hit for one or more fragments, we show if the hit was highly significant (red rectangles, E-value <1e-5), close to the threshold (orange rectangles, ∼1e-5 < E-value < 1), or not significant (yellow rectangles, E-value ≥1). If we could not find a hit for a specific fragment, the corresponding rectangle is gray. The column highlighted between white lines corresponds to the exact hMps1 TPR region. B, secondary structure prediction of the TPR region of hMps1 and the corresponding region in Mps1 orthologues. Alignment and prediction were performed using PSIPRED (33).