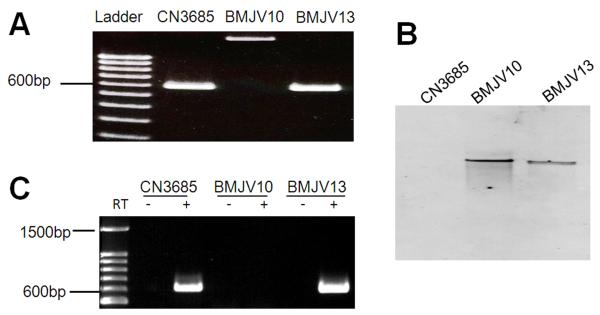
Fig. 1. Inactivation of the agrB gene using the Targetron™ gene knockout system to prepare BMJV10 and complementing strain BMJV13.
Panel A, PCR confirmation of the isogenic agrB null mutant and complementing strain. Using DNA isolated from wild-type CN3685, a PCR assay amplified an ~600 bp product using primers that target the agrB gene. Using DNA purified from the agrB null mutant (BMJV10), which has an ~900 bp intron insertion, this same PCR assay amplified an ~1.5 kb PCR product. Purified DNA from the complementing strain BMJV13 yielded the same ~600 bp PCR product as amplified from the wild-type strain. Lane 1, 100 bp molecular weight marker. Panel B, Southern blot hybridization of a labeled intron-specific probe with DNA from wild-type CN3685, the agrB null mutant (BMJV10) or complementing strain (BMJV13). DNA from each strain was digested with EcoRI and electrophoresed on a 1% agarose gel prior to blotting and hybridization with those above mentioned probes. Panel C, RT-PCR analyses for agrB expression by CN3685, BMJV10 and complementing strain BMJV13 grown overnight in TGY broth. Lane 1 shows size markers. Lanes labeled “+” were from samples receiving reverse transcriptase (RT), while lanes labeled “−“ lacked reverse transcriptase to show the absence of DNA contamination.