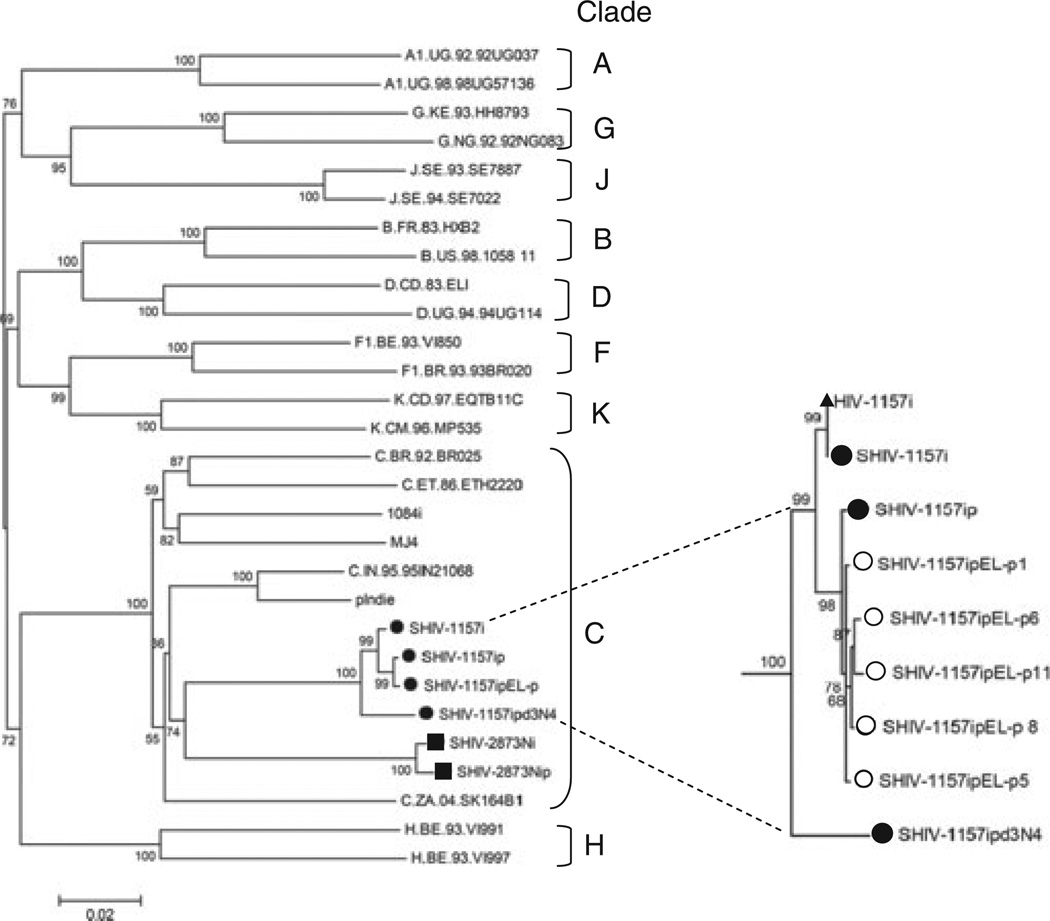
Fig. 2.
Phylogenetic tree construction. Phylogenetic tree showing the relationship between SHIV-1157i, SHIV-1157ip, SHIV-1157ipEL-p, and SHIV-1157ipd3N4 Env sequences and those of other primary strains of HIV. Phylogenetic trees were constructed from full-length Env sequences by using MEGA4. Major clades of HIV group M were used as reference sequences; Env sequences from SHIV-2873Nip and HIV1084i were also included, as SHIV-2873Nip and HIV1084i env were genes derived from the same cohort of HIV-C-infected infants in Lusaka, Zambia. The scale bar indicates the genetic distance along the horizontal branches, and the numbers at the nodes are bootstrap values. The insert represents the phylogenetic relationship between HIV-1157i and the SHIV series related to the env gene of the latter. The open circles denote five individual SHIV-1157ipEL-p sequences obtained through end-point PCR cloning that were used to arrive at the consensus sequence of SHIV-1157ipEL-p. Closed circles represent the consensus sequence of each of the viruses as noted in the figure. The closed triangle represents the parental sequence of HIV-1157i that was used to generate the initial SHIV clone. SHIV, simian–human immunodeficiency virus; HIV-C, HIV clade C.