Fig. 5.
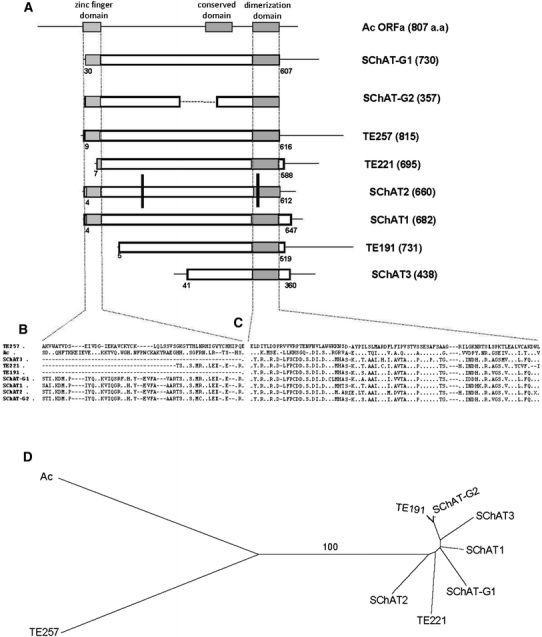
Comparison of hAT transposons recovered from sugarcane genome and the Ac transposase. a Schematic representation comparing the amino acid sequence of the functional transposase of the Ac element from maize, the sugarcane cDNAs TE191, TE221 and TE257, and the predicted amino acid sequences of the transposase of the five SChAT elements found in sugarcane: SChAT1, SChAT2, SChAT3, SChAT-G1 and SChAT-G2. The numbers in brackets in each line correspond to the size of the predicted ORF. The three hAT superfamily domains are highlighted in gray boxes. Black bars indicate the presence of frameshift mutations on the copy of SChAT2. b Detailed alignment of the zinc finger domain. c Detailed alignment of the hAT dimerization domain. d Distance tree based on the concatenated sequences aligned in b and c. Bootstrap value above 50% is presented
