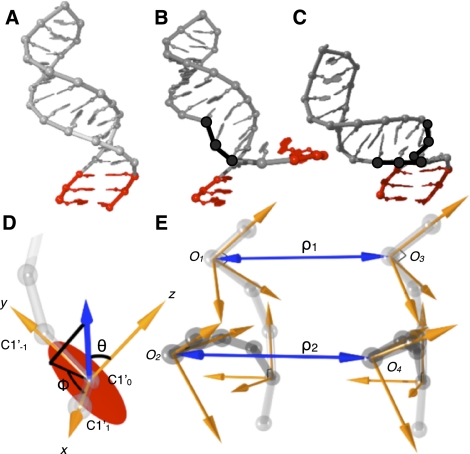
FIGURE 4.
Improved sampling efficiency in RSIM using closed moves. (A) Initial conformation of example RNA molecule. (B) Insertion of random RNA trinucleotide fragment (black) from known high resolution structure library replaces corresponding sequences and disrupts base pairs (red). (C) The closed moves approach replaces pairs of trinucleotide fragments selected to preserve global secondary structure after substitution. (D) To identify appropriate pairs of compensatory RNA fragments, the fragment library is aligned to a local coordinate system with the C1′ atom of first nt being the origin, the C1′ atom of the following nt being along the x-axis, and the C1′ atom of the previous nt defining the positive y-direction. The xy-plane is shown (red circle). The vector defining the relative position of another fragment (blue arrow) sets the coordinate system. (E) Pair of trinucleotide fragments (dark gray) substituted into a prior RNA conformation (light gray). Pairs of trinucleotide fragments are found such that the distance and orientation of connections are preserved. Fragments are first filtered for preservation of distance ρ within 1 Å, followed by filtering for preservation of the relative orientation of the local coordinate systems defined by angles θ and ϕ.